Introduction to Small RNA Sequencing
Small RNAs (sRNAs) are short RNA molecules, usually non-coding, involved with gene silencing and the post-transcriptional regulation of gene expression. sRNA Sequencing (sRNA-seq) is a method that enables the in-depth investigation of these RNAs, in special microRNAs (miRNAs, 18-40nt in length).
Novogene sRNA-seq service is an effective approach to selectively target any species of sRNAs with unprecedented sensitivity and high resolution, all in a single analysis. Coupled with a robust in-house bioinformatics pipeline, Novogene sRNA-seq service has been assisting researchers to describe the differential expression of miRNAs, structural alterations, and to discover novel sRNAs.
Applications of small RNA sequencing
sRNA-seq has been supporting many compelling research avenues, including:
- Profiling known and novel transcripts and expression quantifications of small RNA
- Predicting targeting genes of miRNA candidates
- Identifying tissue, stage, or cell type-specific biomarkers for cancer/disease diagnostics and classification
- Discovering regulatory networks and understanding the mechanisms of tissue or organism development based on transcript profiles
Benefits of Novogene small RNA Sequencing
- The Novogene sRNA-seq is a well-established service that relies on extensive experience, with numerous samples successfully sequenced. It allows access to expression profiles and transcript characterizations for multiple small RNA species, with a tried-and-tested workflow for both input sample of total RNA and exosomal RNA.
- Novogene offers comprehensive analyses using top-notch software and a mature in-house pipeline, meeting all your bioinformatics needs. Researchers can obtain the regulation network, the deep regulation mechanism of gene expression, and more, with Novogene’s high quality services.
sRNA-seq Specifications: RNA Sample Requirements
| Library Type | Sample Type | Amount | RNA Integrity Number (Agilent 2100) | Purity (NanoDrop) |
| Small RNA Library | Total RNA | ≥ 2 μg | Animal ≥ 7.5, Plant ≥ 7, with smooth baseline | OD260/280≥ 2.0; 0D260/230≥ 2.0; no degradation, no contamination |
| Exosomal Small RNA Library | Exosomal RNA | ≥ 10 ng | Fragments between 25-200nt, FU>10 |
Note: Sample amounts are listed for reference only. Download the Service Specifications or Sample Requirements to learn more. For detailed information, please contact us with your customized inquiry.
sRNA-seq Specifications: Sequencing and Analysis
| Sequencing Platform | Illumina NovaSeq 6000 |
| Read Length | Single-end 50 bp |
| Recommended Sequencing Depth | ≥ 10 million read pair per sample |
| Standard Analysis (miRNA) |
|
Note: Sequencing depths and analysis contents displayed are for reference only. Download the Service Specifications to learn more. For detailed information, please contact us.
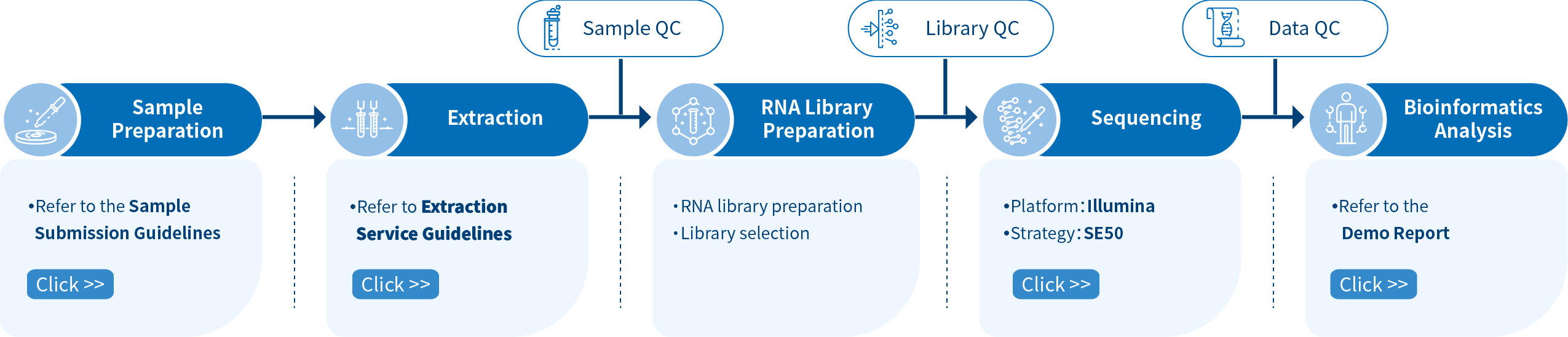
Project Workflow of Novogene sRNA-seq Services
The project workflow starts with sample quality control (Sample QC) to ensure that your samples meet the criteria of the sRNA-Seq technique. Then, the appropriate library is prepared according to your target organism and application, and subsequently tested for its quality (Library QC). Next, a single-end 50 bp sequencing strategy is used to sequence the samples and the resulting data is also checked for its quality (Data QC). Finally, bioinformatic analyses are performed and publication-ready results are provided.
Publications
Novogene’s comprehensive sRNA-seq empowers researchers to investigate the differential expression and regulatory network of miRNAs, structural alterations, and to discover novel small RNAs. Here is a list of publications below that have used Novogene’s sRNA-seq services.
-
A genomic and epigenomic atlas of prostate cancer in Asian populations
NatureIssue Date: 25 March 2020IF: 42.778DOI: 10.1038/s41586-020-2135-x
-
Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana
Nucleic Acids ResearchIssue Date: 19 June 2019IF: 11.501DOI: 10.1093/nar/gkz526
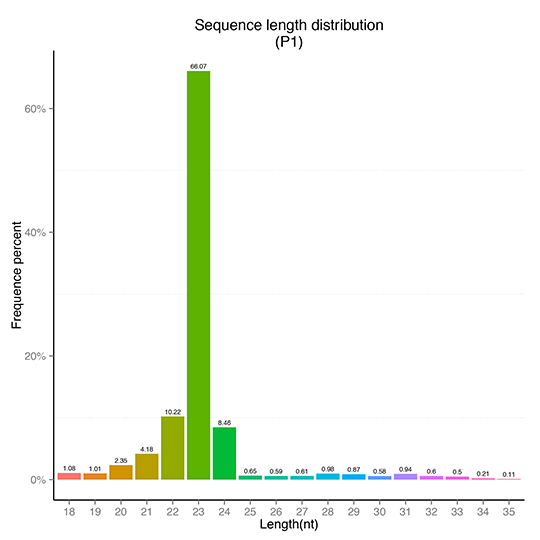
sRNA Length Distribution
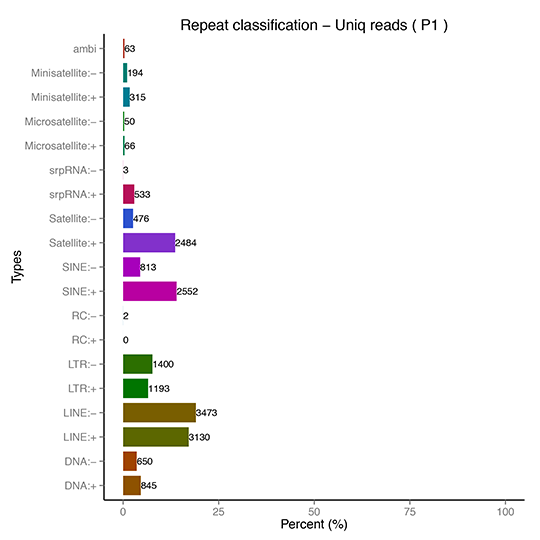
sRNA Classification-Repeat Sequence
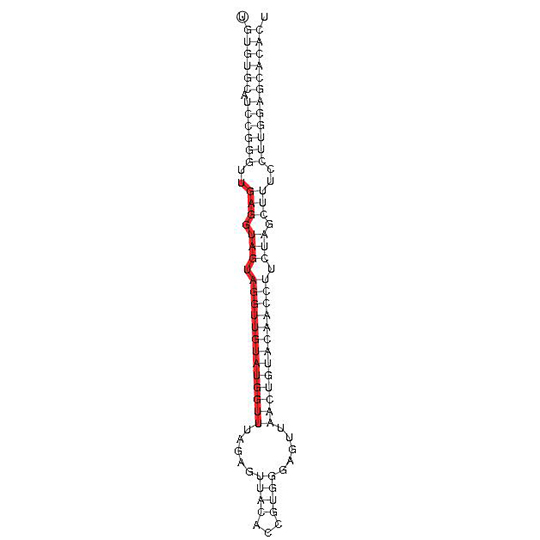
miRNA-structure
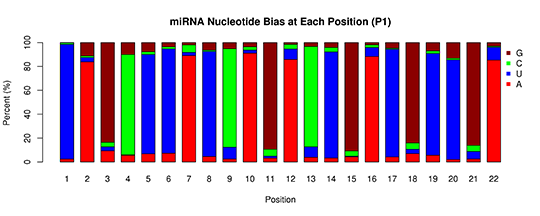
miRNA-base bias
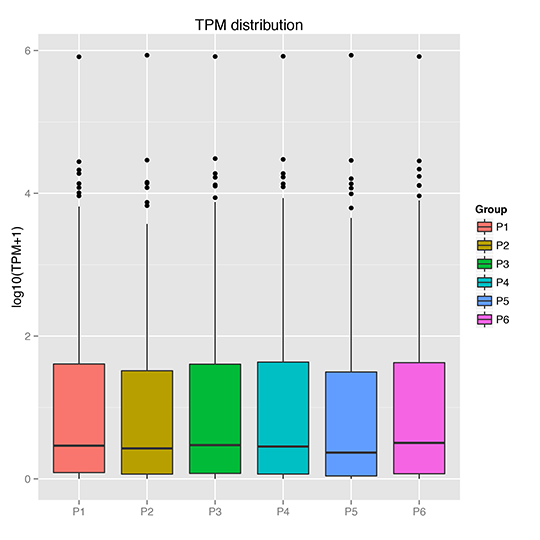
TPM Boxplot
TPM Density Distribution
Hierarchical Cluster
More Research Services