A Beginner’s Guide to Small RNA Sequencing
Ribonucleic acid (RNA) stands as a fundamental constituent in the architecture of life, a molecule essential for comprehending the intricacies of living organisms. RNA sequencing (RNA-seq) is indispensable for researchers seeking to unravel the functional nuances embedded within RNA. Diverse RNA molecules, each with unique roles in regulating gene expression and epigenetic traits, contribute to the dynamic landscape of biological processes. Among these, small RNA (sRNA), classified as a non-coding RNA (ncRNA), has gained prominence by unraveling complex biological mechanisms.
In this blog, you will learn:
- The classification of sRNA
- Library construction and next generation sequencing for sRNAs
- Standard sRNA-seq analysis pipeline
- Applications of sRNA-seq
- How Novogene can help
Keywords: sRNA-seq, small RNAs, non-coding RNAs
Introduction
A distinguishing feature of many types of small RNAs (sRNAs) is their pivotal role in gene silencing, a phenomenon also referred to as RNA silencing. This process is executed through RNA interference (RNAi), where sRNAs actively degrade mRNA, thereby silencing gene expression. The application of sRNA-seq enables researchers to discern the presence of diseases such as breast cancer, Parkinson’s disease, or multiple sclerosis. Additionally, sRNAs contribute to the post-transcriptional regulation of gene expression. Through these multifaceted functions, sRNAs emerge as essential players in the intricate regulatory landscape of cellular processes.
Classification of sRNA
One established method for categorizing non-coding RNAs (ncRNAs) involves consideration of their nucleotide chain length. According to this classification, small RNAs (sRNAs) are characterized as non-coding RNA molecules with a length of fewer than 200 nucleotides. As our understanding of sRNAs has expanded, diverse subtypes of sRNA have been identified, each with unique features and functions:
- Micro-RNA (miRNA): Typically comprising 19 to 25 nucleotides, miRNAs play a crucial role in post-transcriptional regulation by binding to mRNA, thereby influencing gene expression.
- Piwi-interacting RNA (piRNA): Generally ranging from 24 to 31 nucleotides, piRNAs are predominantly associated with protecting the genome from transposons, contributing to genomic stability, particularly in germ cells.
- Small interfering RNA (siRNA): A less common double-stranded RNA molecule, typically spanning 20 to 25 base pairs. siRNAs are central players in RNA interference (RNAi), actively participating in the degradation of target mRNA molecules, thereby silencing gene expression.
- Small nuclear RNA (snRNA): Typically falling within the range of 100 to 150 nucleotides, snRNAs are integral components of small nuclear ribonucleoproteins (snRNPs), contributing to the splicing of pre-mRNA during transcription.
- Small nucleolar RNA (snoRNA): Typically exhibiting lengths between 60 to 170 nucleotides, snoRNAs are primarily involved in the modification and processing of ribosomal RNA (rRNA) and transfer RNA (tRNA).
Next Generation Sequencing for sRNAs
The key breakthrough in advancing the understanding of small RNAs (sRNAs) was achieved through the implementation of next-generation sequencing (NGS). This technology prioritizes ultra-high throughput, scalability, and rapidity in determining nucleotide sequences of RNA samples. In comparison to preceding methods like microarrays, NGS demonstrates superior capabilities for the detection of novel RNA transcripts, delivering expedited results with heightened specificity and sensitivity.
Sequencing sRNA using NGS is a complex process that unfolds over four stages. Let’s look at each of them in greater detail:
- RNA sample collection: The process commences with the extraction of RNA from targeted cells, ensuring sample quality through post-extraction RNA enrichment.
- RNA library preparation: Once the sRNA samples are ready, a library of cDNA is created. This is accomplished through reverse transcription of RNA molecules to derive cDNA. The fact that sRNA molecules do not need to be fragmented before library preparation sets sequencing apart from other types of ncRNA-seq.
- cDNA sequencing: The cDNA library undergoes sequencing via powerful NGS platforms, employing a single-end 50 bp sequencing strategy for comprehensive data acquisition.
- Bioinformatic analysis: Upon successful completion of sequencing and quality checks, the obtained data undergoes rigorous bioinformatic analysis. We dive into the intricacies of one such sRNA-seq pipeline in our next section.
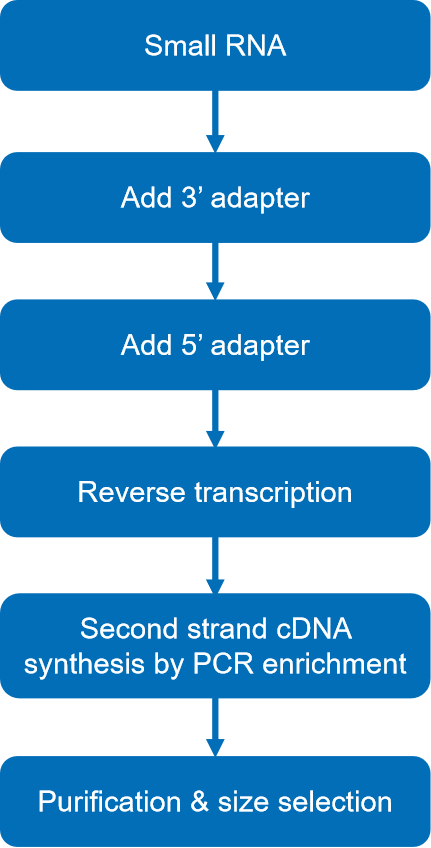
Fig 1. Library construction workflow
Standard sRNA-seq Analysis Pipeline
The standard pipeline for sRNA-seq analysis, applicable across various sRNA types such as miRNA, piRNA, and siRNA, unfolds as follows:
- Raw data collection and quality control: Initial data collection followed by a quality control check.
- Length filtering and mapping: RNA molecules undergo length filtering, mapping the distribution of sample molecule lengths. The identification of sRNA molecules as common or specific also occurs during this stage.
- Genome mapping: Mapping of sRNA molecules to the reference genome of the organism.
- Classification and quantification: Grouping of sRNAs into clusters based on type, including known miRNA, rRNA, tRNA, snRNA, snoRNA, repeat-associated sRNA, exon, intron, and predicted novel miRNA.
- Analysis of known and novel miRNAs: Detailed analysis of both known and predicted novel miRNAs.
- Enrichment analysis of target genes: Exploration of differentially expressed miRNAs through enrichment analysis of target genes.
The table below illustrates the entire pipeline:
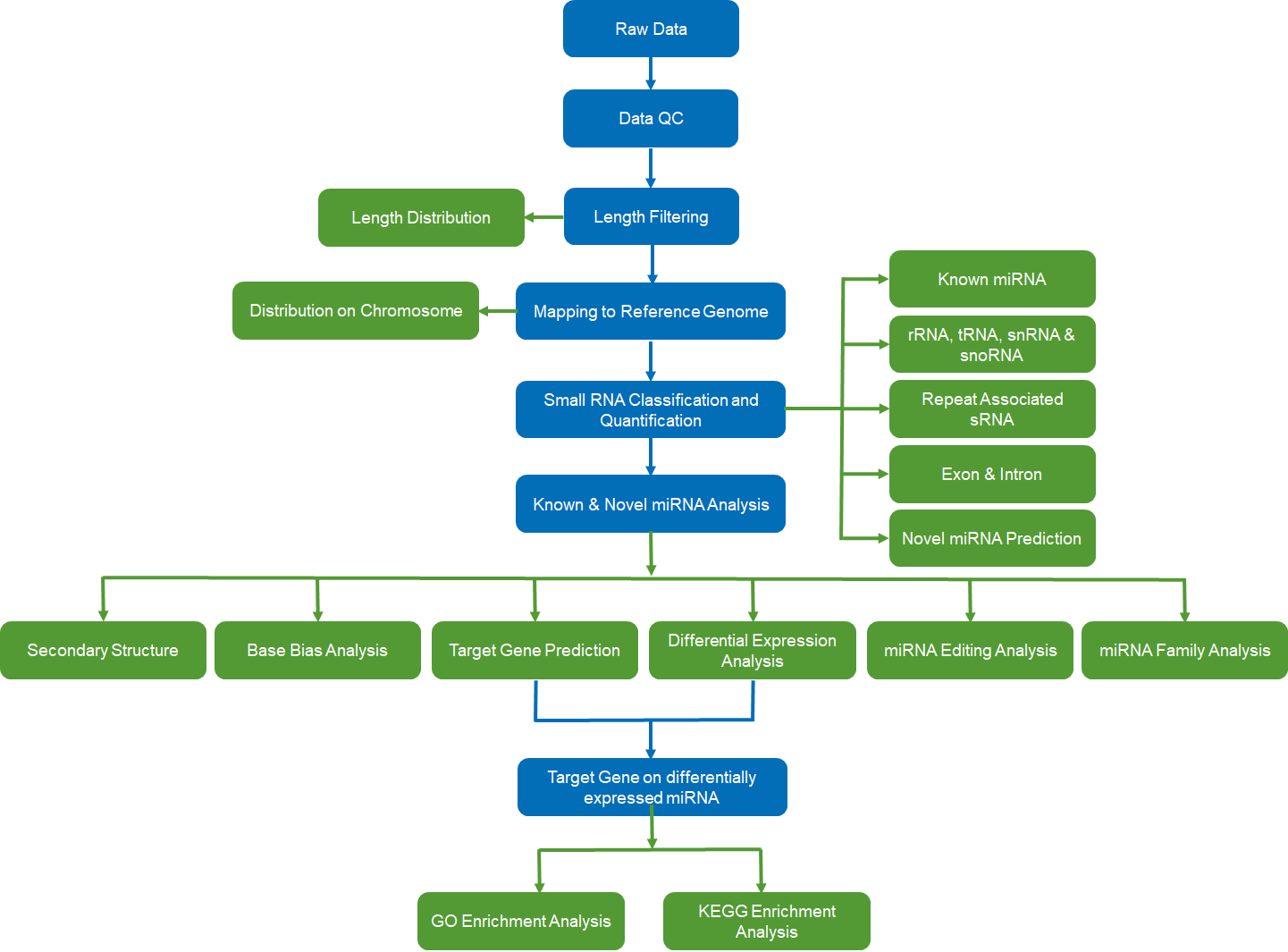
Fig 2. Bioinformatics analysis pipeline
Applications of sRNA-Seq
Small RNA sequencing (sRNA-Seq) serves as a robust research tool with diverse applications:
- Profiling known and novel sRNA transcripts: Systematic identification and characterization of both established and novel small RNA transcripts within biological samples.
- Express quantification of sRNA: Accurate measurement of small RNA expression levels,facilitating quantitative analyses in various biological contexts.
- Predicting miRNA targeted genes: Computational prediction and identification of target genes regulated by microRNAs (miRNAs), offering insights into post-transcriptional gene regulation.
- Identifying tissue, stage, or cell type-specific biomarkers for disease: Exploration of sRNA profiles to discern specific biomarkers associated with diseases in various tissues, developmental stages, or cell types.
- Discovering regulatory networks for tissue or organism development: Unraveling regulatory networks based on small RNA transcript profiles, providing a deeper understanding of tissue or organism development dynamics.
The applications outlined above underscore the versatility and utility of sRNA-seq in elucidating functional aspects of small RNA molecules across diverse biological contexts.
One particular use of sRNA-seq in the case of miRNAs we would highlight was performed by Li et al. (2023). The authors used a combination of miRNA sequencing (miRNA-Seq) and bioinformatics analysis to identify miRNAs associated with colorectal cancer and to investigate the role of miRNAs in post-transcriptional control. The authors combined the expression profiles obtained from in-house miRNA-seq, TCGA, GEO, and other array databases from the National Center for Biotechnology Information (NCBI) web server to identify miRNAs associated with colorectal cancer. They then used bioinformatics analysis to predict the target genes of the identified miRNAs and to investigate the functional implications of miRNA binding on gene expression. They also performed luciferase reporter assays to validate the predicted miRNA binding sites in the 3’-UTR of BET1L and other target genes. These techniques allowed the researchers to identify miR-140-3p as a key regulator of BET1L expression and to investigate the functional implications of miRNA binding on gene expression.
How Novogene Can Help
Using a single-end strategy of the Illumina NovaSeq sequencing platform, Novogene’s sRNA-seq services deliver comprehensive, reliable results. This NGS method allows Novogene to accurately capture the entire sRNA transcriptome with extreme sensitivity and high resolution in a single analysis. Have all your sRNA bioinformatics requirements met with the help of Novogene’s sRNA-Seq services.
Links & References
- https://www.novogene.com/us-en/services/research-services/transcriptome-sequencing/non-coding-rna-sequencing/small-rna-sequencing-srna-seq/
- https://www.novogene.com/eu-en/resources/onlineevent/a-beginners-guide-to-rna-seq-advanced/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4196292/
- https://pubmed.ncbi.nlm.nih.gov/25747396/
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7152241/
- https://sapac.illumina.com/techniques/sequencing/rna-sequencing/small-rna-seq.html
- https://aacrjournals.org/cancerres/article-abstract/83/13/2142/727428/Genetic-Modulation-of-BET1L-Confers-Colorectal?redirectedFrom=fulltext
