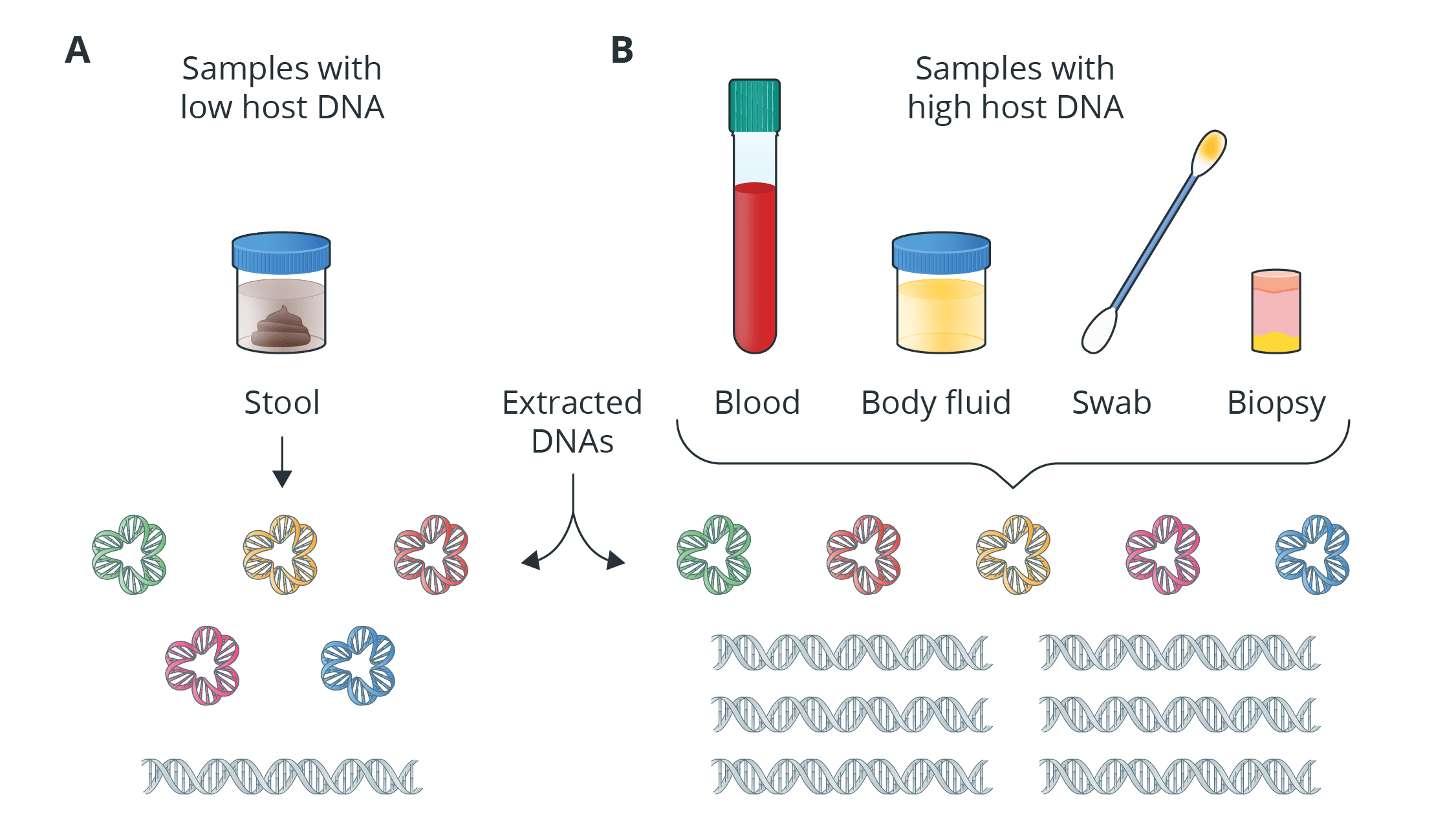
Introduction to Shotgun Metagenomic Sequencing
Shotgun metagenomic sequencing provides information on the total genomic DNA from all organisms in a sample, which is called metagenome, and there is no need to isolate and cultivate the microorganisms or amplification of target regions. This is crucial during the study of metagenome because it is believed that nearly 99% of all microorganisms cannot be cultivated in the laboratory. Unlike the targeted approach used in the 16S/18S/ITS amplicon sequencing, shotgun metagenomic sequencing provides not only information on the taxonomic annotations of each organism, but also the functional profiling, gene prediction and microbial interaction of the whole community, through shotgun sequencing approach.
With extensive experience and well-developed bioinformatics pipelines, our Metagenomic Shotgun sequencing services can help you with your metagenomics study, including determining the genomes, functions, and variations of microorganisms from diverse samples. Novogene’s Metagenomic shotgun sequencing service delivers high-quality data, publication-ready results, and personalized analysis to meet different research objectives. Novogene offers comprehensive metagenomics solutions, from the study of community structure and species classification to system evolution, gene function, and metabolic network of environmental microorganisms.
Applications of Shotgun Metagenomic Sequencing
From taxonomic profiling to functional analysis and metabolic networks, shotgun metagenomic sequencing can be used in many different research topics, including:
- Annotation of microbial composition and function
- Detection of microbes of interest related to human health, environmental recovery, and energy synthesis
- Deep investigation of the genetic makeup and metabolism of microbes for drug development
- Investigation of the microbe-host relationship, which can also be related to the development of new drugs
Benefits of Novogene Shotgun Metagenomic Sequencing
- Shotgun metagenomic sequencing can be used to sequence multiple microorganisms, all in a single run, without specific isolation and cultivation of microorganisms or the amplification of target regions for either prokaryotes or eukaryotes. The results of metagenomic sequencing provide complete information not only about microbial diversity but also functional diversity and metabolic pathways in the samples.
- Novogene has extensive experience providing metagenomic sequencing services to help clients obtain information from many different sample types, including soil, fecal, and water. Our newly improved metagenome assembly pipeline gives us the ability to provide long-read metagenome assembly services (PacBio Sequel II platform). This exciting development has made it possible to achieve complete maps of certain bacteria and fungi for taxonomy resolution and functional annotation.
Metagenomic Specifications: DNA Sample Requirements
| Sample Type | Amount | Volume | Concentration | Purity |
| Total DNA | ≥ 200 ng | ≥20 μL | ≥ 10 ng/μL | A260/280 = 1.8-2.0 |
Note: Sample amounts displayed are for reference only. Download the Service Specifications to learn more. For detailed information, please contact us with your customized requests.
Metagenomic Specifications: Sequencing and Analysis
| Sequencing Platform | Illumina NovaSeq X Plus |
| Read Length | Pair-end 150 bp |
| Recommended Sequencing Depth | ≥ 20 million read pairs (6Gb data) per sample for standard analysis ≥ 10 million read pairs (3Gb data) per sample for Reads Mapping analysis Depending on analysis contents and host sequence ratio. |
| Standard Data Analysis (Extensive analysis contents) |
|
| Reads Mapping Analysis (Cost-effective and rapid) |
Two optional pipelines: MetaPhlAn and Kraken
|
Note: Sequencing depths and analysis contents listed are for reference only. Download the Service Specifications to learn more. For detailed information, please contact us.
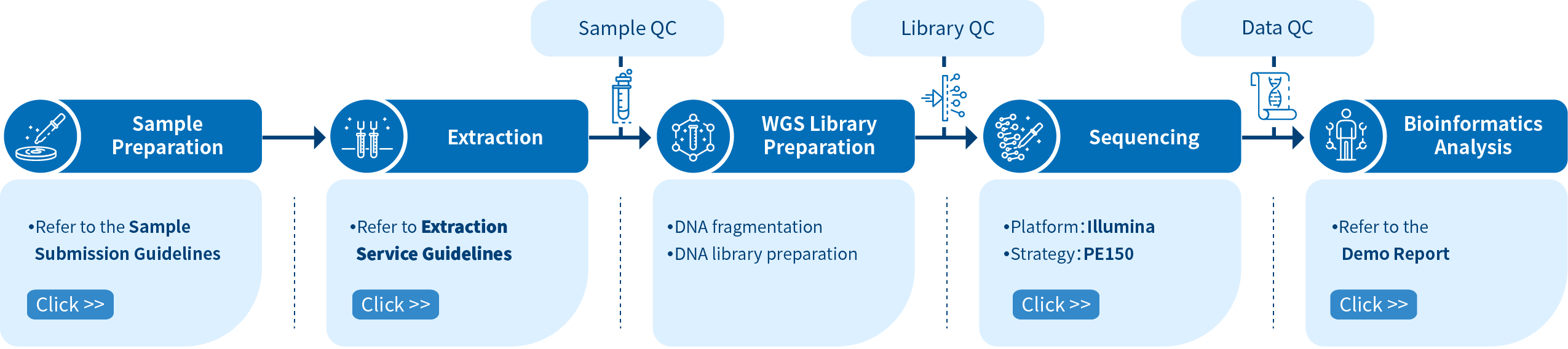
Project Workflow of Novogene Metagenomic Sequencing Services
In order to ensure the accuracy and reliability of the sequencing data, quality control (QC), consistent with our high scientific standards, is performed at each step of the procedure. The shotgun metagenomic sequencing workflow comprises these main steps: sample preparation and quantification, fragmentation and library preparation, library quality control, sequencing, and bioinformatics analysis.
Featured publications with Novogene Shotgun Metagenomic Sequencing service
Shotgun Metagenomic Sequencing is a flexible approach to obtain microbial genomes among environments (de novo assembly), as well as gene functions (function annotation), and how they interact with each other (pathway analysis). Here presents a list of academic publications that have used Novogene Shotgun Metagenomic Sequencing services.
-
Effects of spaceflight on the composition and function of the human gut microbiota
Gut MicrobesIssue Date: 2020 Jul 3IF: 7.740DOI: 10.1080/19490976.2019.1710091
-
MicrobiomeIssue Date: 19 October 2018IF: 11.607DOI: 10.1186/s40168-018-0561-x
-
Hypoxia induces senescence of bone marrow mesenchymal stem cells via altered gut microbiota
Nature CommunicationsIssue Date: 2018 May 22IF: 12.121DOI: 10.1038/s41467-018-04453-9
-
Age-associated microbiome shows the giant panda lives on hemicelluloses, not on cellulose
ISME JournalIssue Date: 01 February 2018IF: 9.180DOI: 10.1038/s41396-018-0051-y
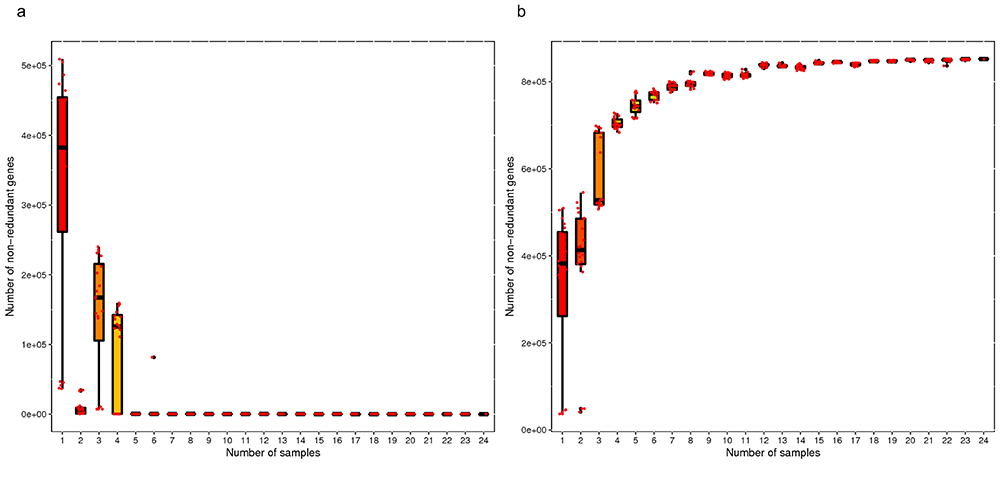
Core-pan Gene Rarefaction Curve
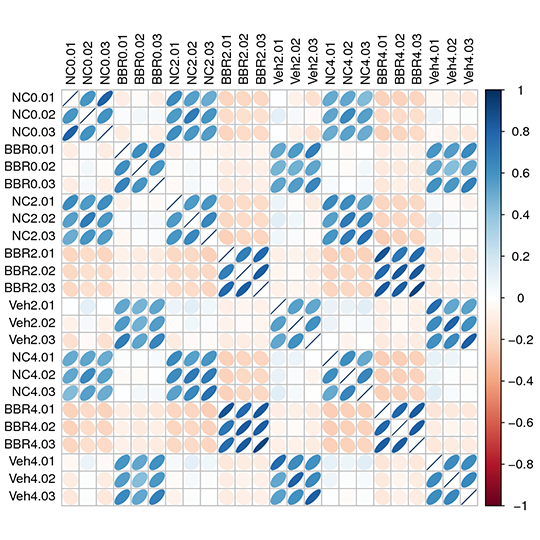
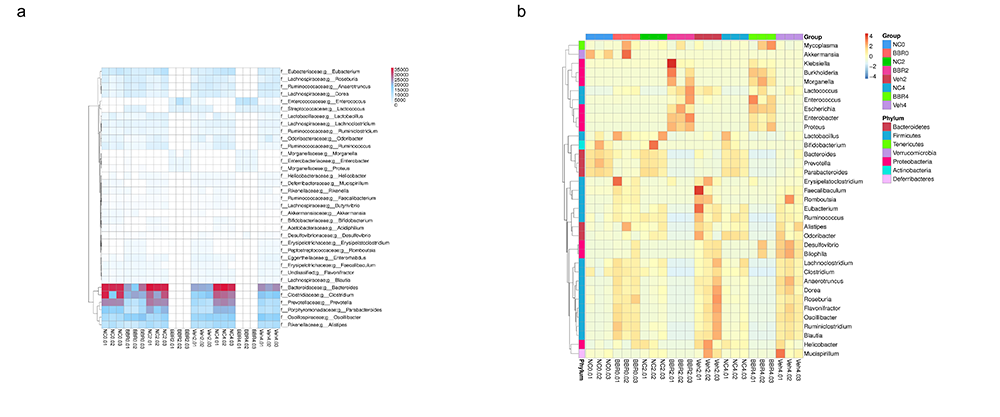
Heatmap for the Relation among samples
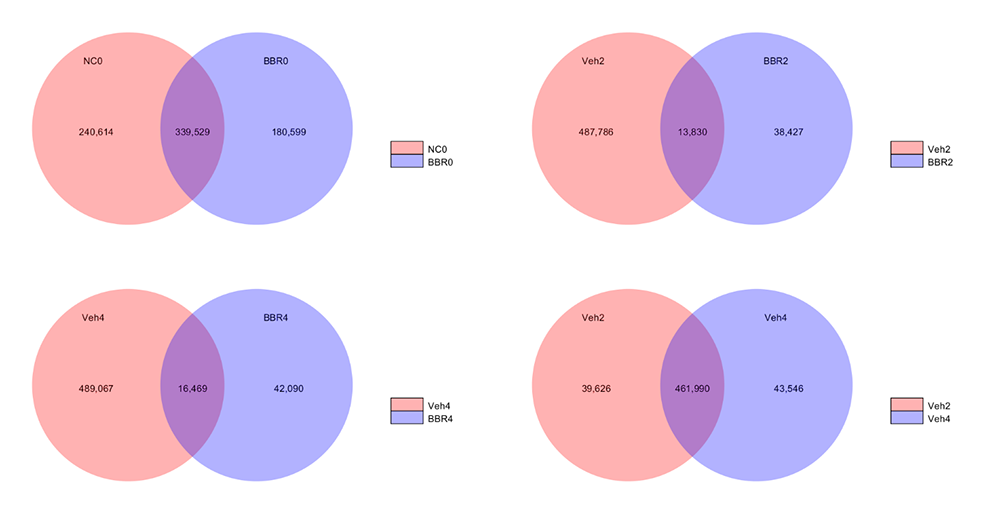
Venn Figures for Gene Number among samples
Gene Number & Abundance Clustering Heatmap in genus level
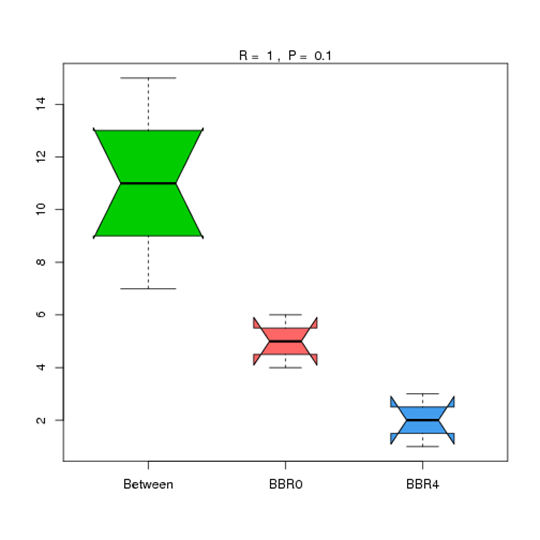
Anosim Analysis at Phylum Level
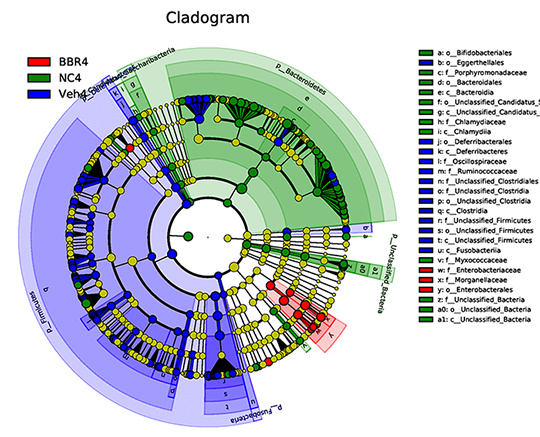
Linear Discriminant Analysis (LDA) & Cladogram
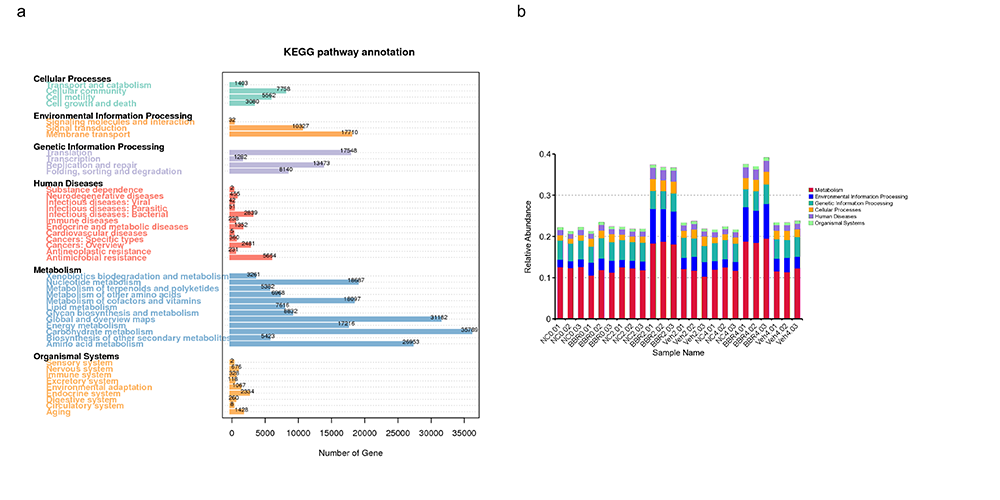
Kyoto Encyclopedia of Genes and Genomes (KEGG) Annotation
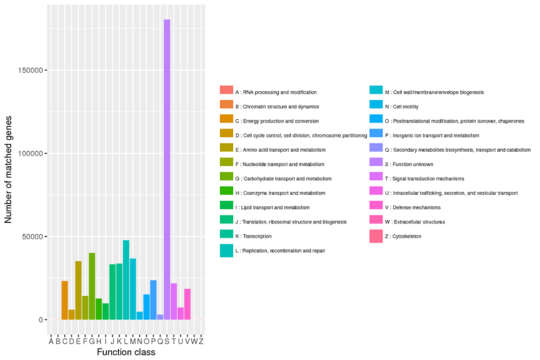
Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups (eggNOG) Annotation
PCA & NMDS Analysis Results based on relative abundance of gene functions

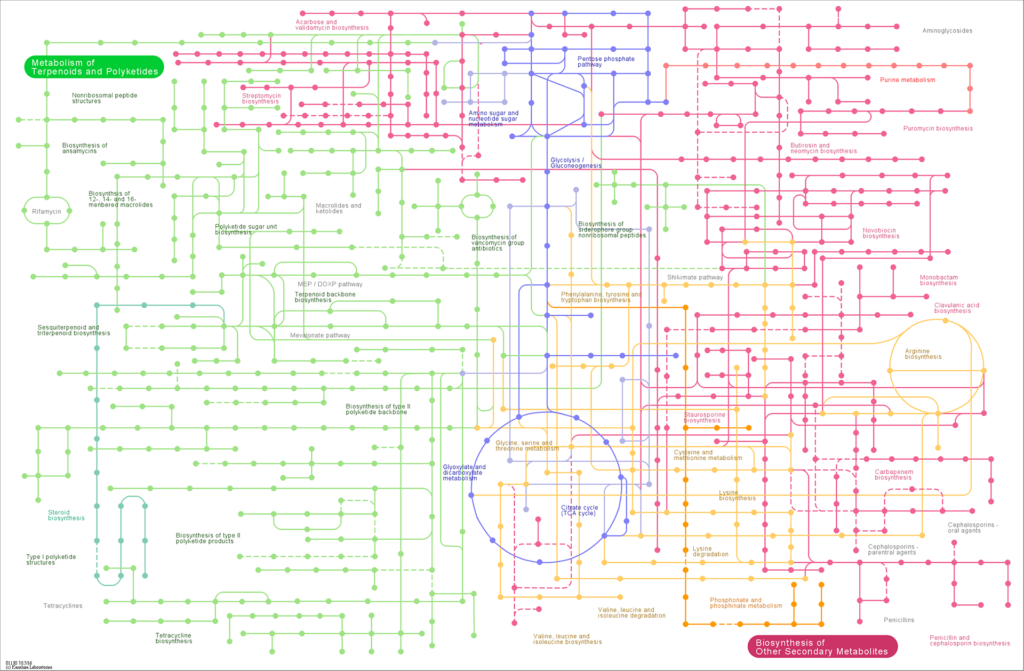
Comparison Plot for Metabolic Pathway of multi-samples
More Research Services