Introduction to Whole Genome Bisulfite Sequencing
DNA methylation at the C5 position of cytosine plays a crucial role in gene expression and chromatin remodeling. Perturbations in methylation patterns are associated with tumorigenesis, neurodegenerative diseases and neurological disorders. Bioinformatic analysis of methylomes is widely used in various research areas, including studies on gene regulation, cell differentiation, embryogenesis, aging, occurrence and development of disease, phenotypic diversity and evolution in plants and animals.
Novogene provides whole genome bisulfite sequencing (WGBS) to efficiently identify methylated cytosines on a genome-wide scale with a single nucleotide resolution. The deep reach of the methylome is imperative for understanding gene expression and multiple biological processes that are subject to epigenetic regulation.
Applications of Whole Genome Bisulfite Sequencing
Novogene delivers high-quality data and publication-ready analysis results of WGBS services, to faciliate research including:
- Profiling methylation patterns at CG, CHG and CHH sites with single nucleotide resolution
- Identifying differentially methylated sites associated with experimental treatments or sample conditions
- Understanding mechanisms of cell differentiation or tissue development on the basis of methylation profiles
- Early diagnosis of diseases and cancers through the detection of DNA hypermethylation or hypomethylation
Benefits of Novogene WGBS Service
- Hundreds of projects have been successfully completed with highly professional, publication-quality data output and bioinformatics analysis.
- Ultra-low input DNA and high bisulfite conversion rate are available in methylation library preparation.
- Industry-standard software (such as Bismark) and mature in-house pipelines are used for mapping and comprehensive bioinformatics analysis.
- Customized association analysis can explore correlations between methylation and gene expression.
WGBS Specifications: DNA Sample Requirements
| Sample Type | Required Amount | Purity |
| Genomic DNA | ≥ 100 ng | A260/280=1.8-2.0 |
Note: Sample amounts are listed for reference only. Download the Service Specifications to learn more. For detailed information, please contact us with your customized requests.
WGBS Specifications: Sequencing and Analysis
| Platform Type | Illumina NovaSeq 6000 |
| Read Length | Paired-end 150 bp |
| Sequencing Depth | ≥ 30× coverage for the species with reference genome |
| Standard Data Analysis |
|
| WGBS mRNA-seq Co-Analysis (Integrated Analysis of Methylome and Transcriptome) |
|
Note: Sequencing parameters and analysis contents displayed are only for reference. Download the Service Specifications to learn more. For detailed information, please contact us with your customized requests.
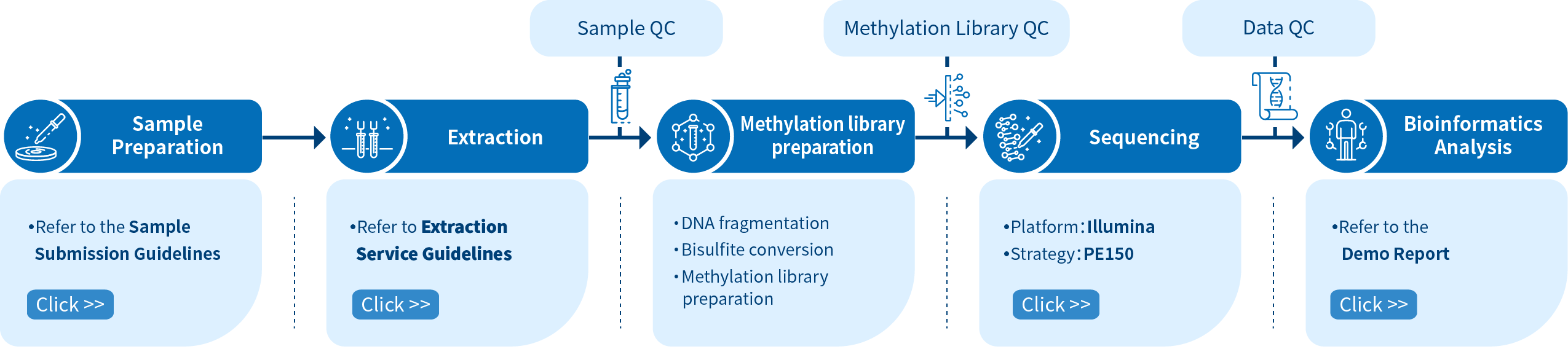
Project Workflow of Novogene WGBS Service
The Novogene WGBS service is comprised of four steps. Sample preparation is followed by library preparation, sequencing and bioinformatics analysis. To construct a methylation library, DNA fragmentation is performed initially, and sodium bisulfite converts the unmethylated cytosines into uracils. Libraries are then sequenced using Illumina PE150 and the reads are processed with bioinformatics pipelines.
In order to ensure the accuracy and reliability of sequencing data, Novogene audits every experimental step strictly by quality control and ensures high-quality data output fundamentally, from DNA sampling to obtaining the final data report. Obtaining high-quality data is the premise to ensure that bioinformatics analysis is correct, comprehensive and credible.
Featured Publications using Novogene WGBS Service
Whole Genome Bisulfite Sequencing (WGBS) is a powerful method to identify individually methylated cytosines on a genome-wide scale. The deep reach of the methylome (including methylated cytosine detection), the promoter detection of differentially methylated sites and regions, and related functional analysis of the Differentially Methylated Region (DMR) and Differentially Methylated Promoter (DMP), are all essential for understanding gene expression and other processes that are subject to epigenetic regulation. Here summarizes a list of publications that used Novogene WGBS service.
-
PRC2-AgeIndex as a universal biomarker of aging and rejuvenation
Nature CommunicationsIssue Date: 16 July 2024IF: 14.7DOI: 10.1038/s41467-024-50098-2
-
Mutant IDH1 inhibition induces dsDNA sensing to activate tumor immunity
Science Issue Date: 6 July 2024IF: 44.7DOI: 10.1126/science.adl6173
-
Nucleic Acids Research Issue Date: 5 July 2024IF: 16.6DOI: 10.1093/nar/gkae572
-
Nature CommunicationsIssue Date: 25 May 2024IF: 14.7DOI: 10.1038/s41467-024-48940-8
-
Auto-suppression of Tet dioxygenases protects the mouse oocyte genome from oxidative demethylation
Nature Structural & Molecular BiologyIssue Date: 4 January 2024IF: 16.8DOI: 10.1038/s41594-023-01125-1
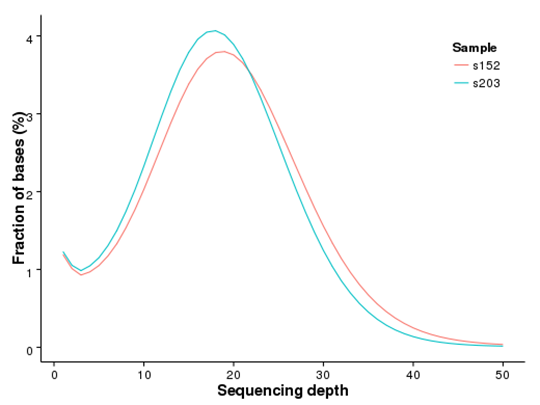
Distribution of Genome Coverage
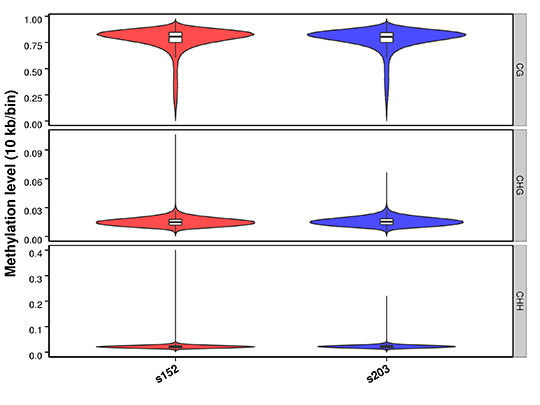
Methylation Level Distribution on Whole Genome
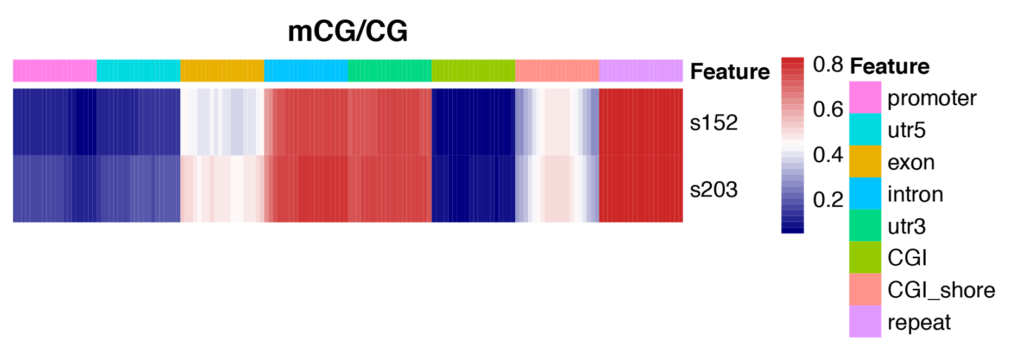
Heatmap Analysis for Methylation Levels of Gene Functional Region
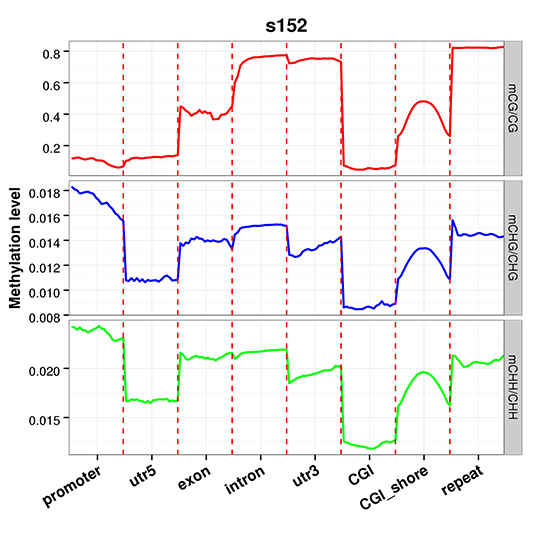
Methylation Level Distribution at Functional Genetic Elements
Circos Plot for DMR Condition in three contexts (CG, CHG, CHH)
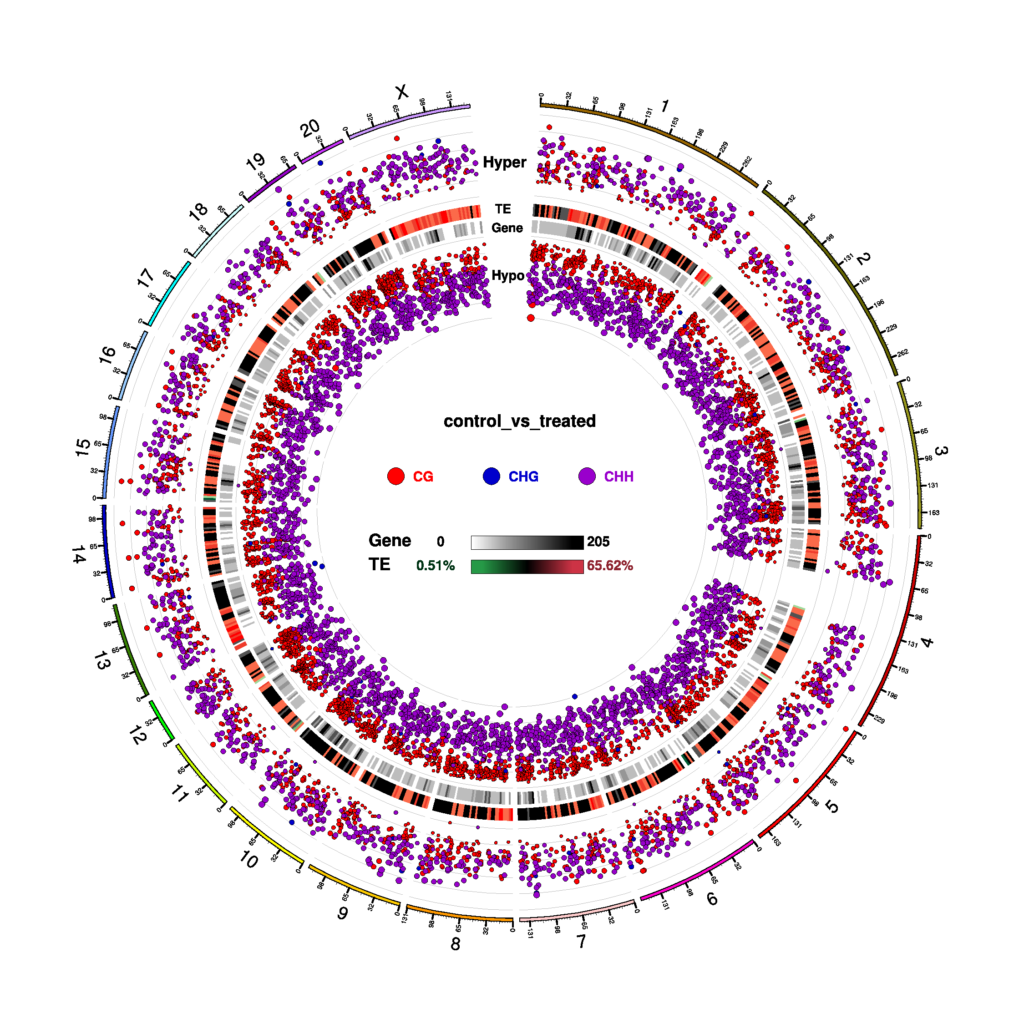
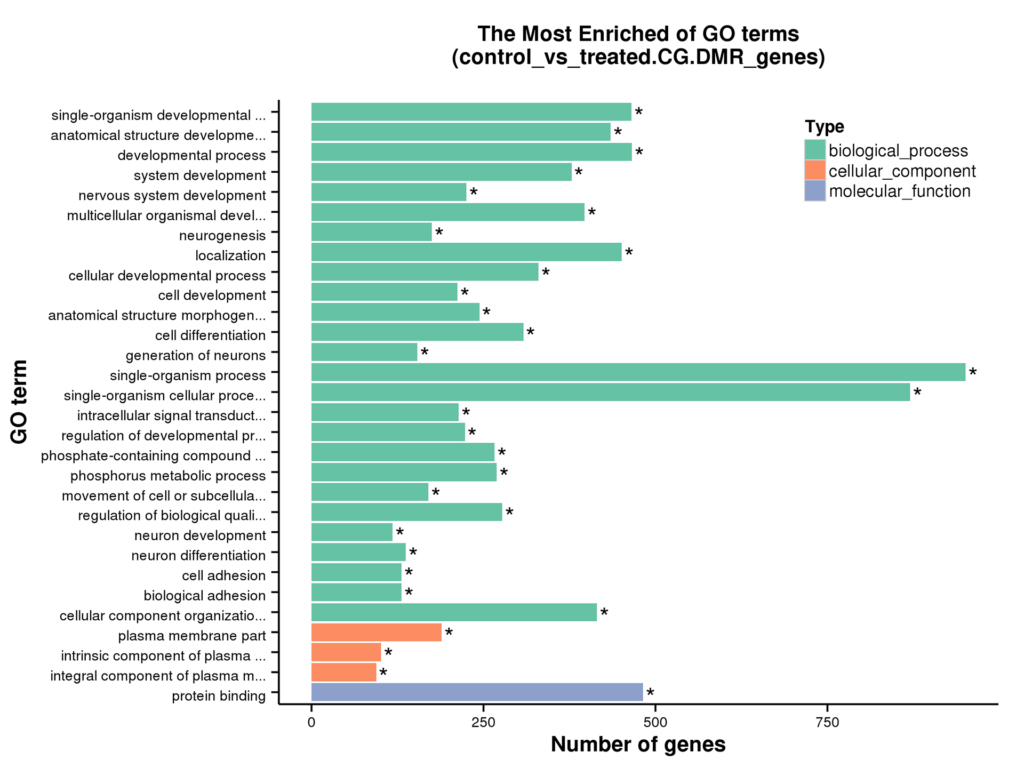
Diagram of GO Enrichment