Introduction to lncRNA Sequencing
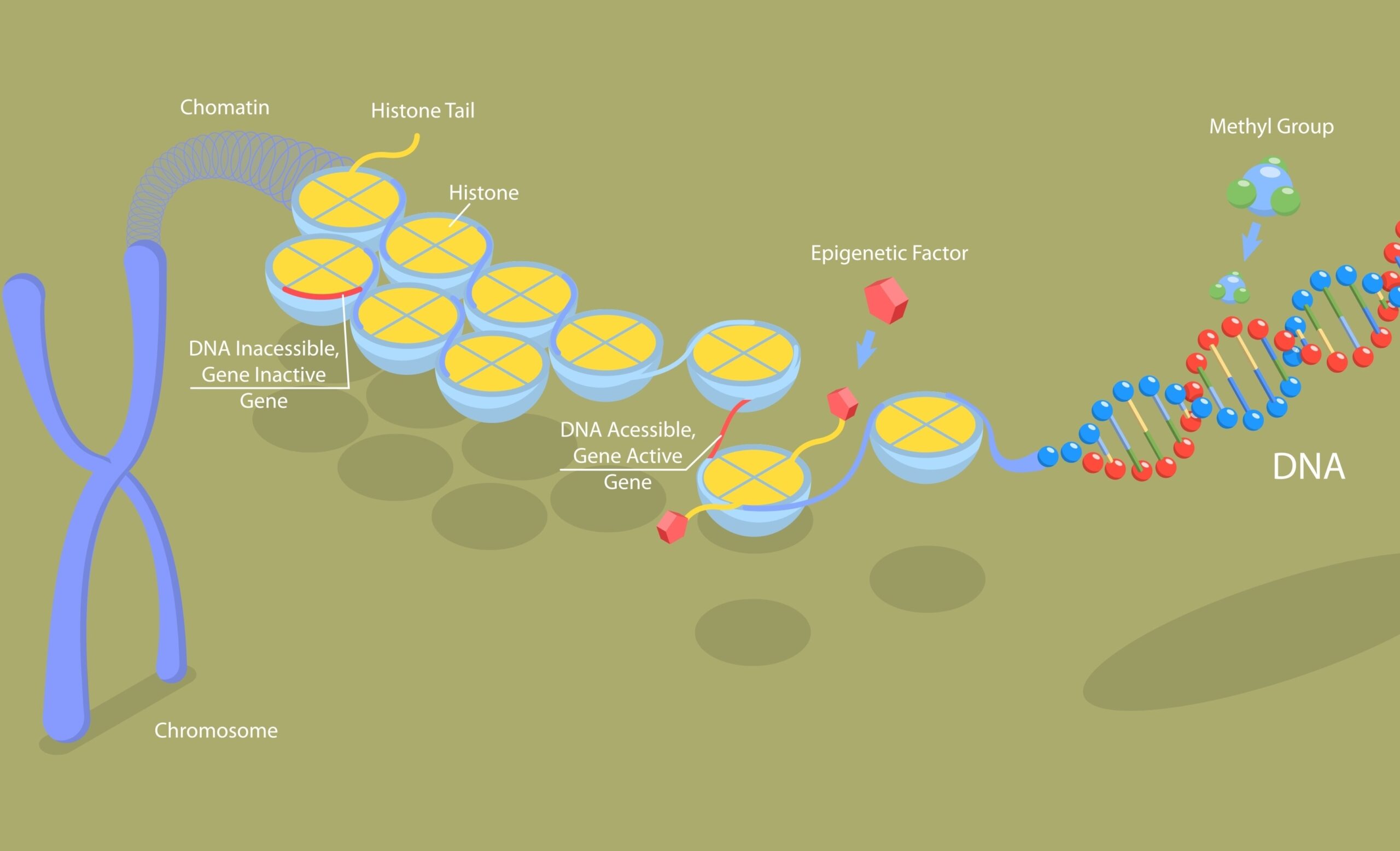
Long non-coding RNAs (lncRNAs) are a moderately abundant fraction of the eukaryotic transcriptome, which are comprised of longer than 200nt non-coding RNAs (ncRNAs) , including lincRNAs (intergenic lncRNAs), intronic, anti-sense, sense and bidirectional lncRNAs, which do not encode proteins. lncRNAs affect multiple cellular functions through their regulation of gene transcription, post-transcriptional modifications, and epigenetics. lncRNA sequencing (lncRNA-seq) is a powerful NGS tool to study functional roles in diverse biological processes and human diseases, such as cancer and neurological disorders.
The progressive library preparation of Novogene’s lncRNA-seq service enables information enrichment and gene expression profiling for both coding and non-coding transcripts from a highly sensitive transcriptomic perspective. Through the application of bioinformatics analysis, the strand orientation and the regulatory association between lncRNA and targeted mRNA can be investigated, all in a single run.
Applications of lncRNA sequencing
LncRNA-seq has been opening new and exciting research possibilities, including:
- Profiling known and novel transcripts and identifies variations
- Predicting target genes of lncRNA
- Identifying biomarkers for cancer/disease diagnostics and classification
- Discovering regulation between lncRNA and mRNA
Benefits of Novogene lncRNA sequencing
- Novogene lncRNA-seq offers high throughput and high accuracy (with Q30 score ≥ 85%) coupled to low initial RNA amounts required. Novogene has extensive experience providing RNA-Seq services, having successfully completed thousands of projects and helped multiple researchers to publish in high Impact Factor journals.
- Novogene offers inclusive solutions for lncRNA identification, quantification, differential expression, enrichment analysis, new gene prediction and target prediction. Highly-qualified bioinformaticians deliver publication-ready analysis results using personalized pipelines.
lncRNA-seq Specifications: RNA Sample Requirements
| Library Type | Sample Type | Amount | RNA Integrity Number (Agilent 2100) | Purity (NanoDrop) |
| lncRNA Library (rRNA depletion) |
Total RNA | ≥ 500 ng | ≥ 5.5, with smooth baseline |
OD260/280≥ 2.0; 0D260/230≥ 2.0; no degradation, no contamination |
| Ultra-low input total RNA directional library | Ultra-low total RNA(human, mouse, rat) | ≥ 25ng | ≥5.5, with flat baseline | |
| Ultra-low input total RNA directional library | Ultra-low total RNA(blood) (human, mouse, rat) | ≥ 120ng | ≥5.5, with flat baseline | |
| Exosomal lncRNA Library | Exosomal RNA | ≥ 5 ng | Fragments between 25-200nt, FU*>10 |
|
| Dual RNA library | Total RNA | ≥ 1 μg | ≥ 6.5, with flat base line |
Note: Sample amounts displayed are for reference only. Download the Service Specifications to learn more. For detailed information, please contact us.
lncRNA-seq Specifications: Sequencing and Analysis
| Sequencing Platform | Illumina NovaSeq 6000 |
| Recommended Data Output | ≥ 40 million read pairs per sample |
| Recommended Sequencing Depth | ≥ 40 million read pairs per sample for species with reference genome |
| Standard Data Analysis |
|
Note: Parameters are displayed for reference only. Download the Service Specifications to learn more. For detailed information, please contact us.
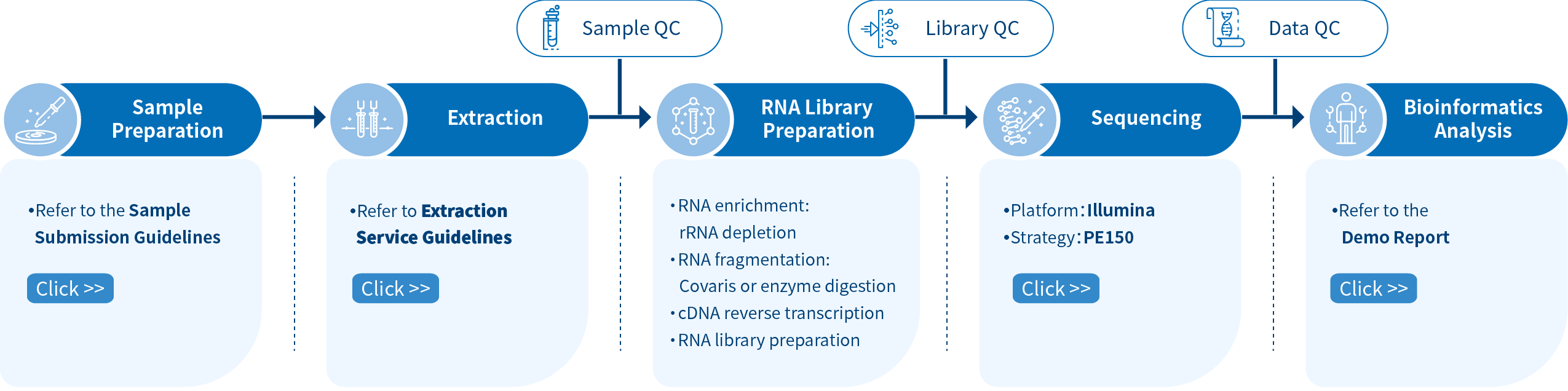
Novogene lncRNA Sequencing Project Workflow
The workflow of the lncRNA-seq starts with sample preparation and quality control. The ribosomal RNA (rRNA) is depleted for target transcript enrichment. The fragmented RNA undergoes reverse transcription into cDNA. Strand-specific libraries (also known as stranded libraries, directional libraries) are prepared, and the sequencing is performed using a paired-end 150bp strategy on the Illumina platform. The downstream processing follows the well-established Novogene pipeline, which guarantees the highest quality results. Customized bioinformatics solutions are available upon request.
Featured publications using Novogene lncRNA service
lncRNA-seq is a powerful tool that not only reveals the quantification and functional enrichment of the target transcripts but also indicates the strand orientation and regulatory relationship between lncRNA and targeted mRNA. There list some exciting publications that have used Novogene’s lncRNA-seq services.
-
Celll ReportsIssue Date: 2020IF: 8.109DOI: 10.1016/j.celrep.2020.107694
-
Small Protein Hidden in lncRNA LOC90024 Promotes “Cancerous” RNA Splicing and Tumorigenesis
Advanced ScienceIssue Date: 11 March 2020IF: 15.840DOI: 10.1002/advs.201903233
-
Molecular PsychiatryIssue Date: 03 February 2020IF: 12.384DOI: 10.1038/s41380-020-0662-3
-
Plant Biotechnology JournalIssue Date: 16 August 2019IF: 8.154DOI: 10.1111/pbi.13234
-
rSj16 Protects against DSS-Induced Colitis by Inhibiting the PPAR-α Signaling Pathway
TheranosticsIssue Date: 2017IF: 8.579DOI: 10.7150/thno.20359
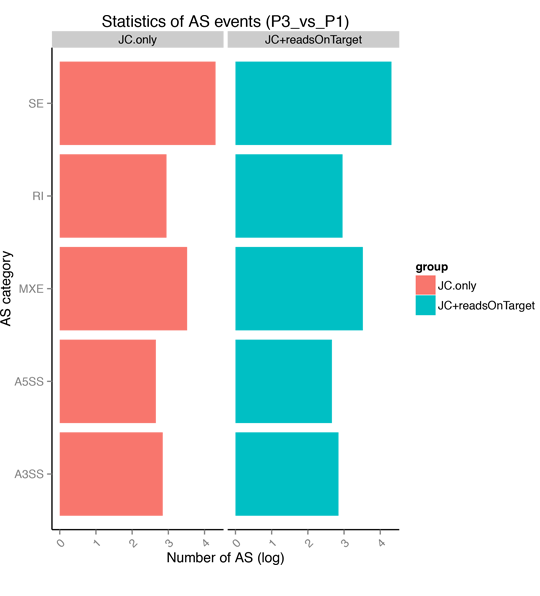
Alternative Splicing
lncRNA Identification
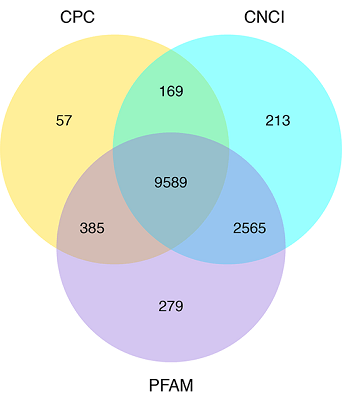
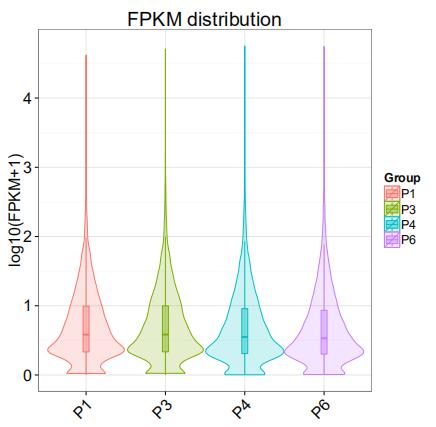
Group Comparison
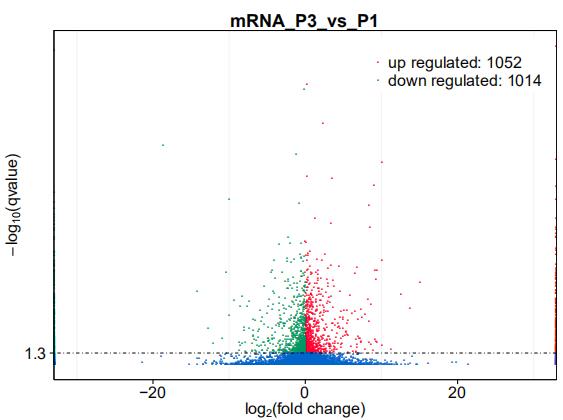
Volcano Plot of Transcript Quantification
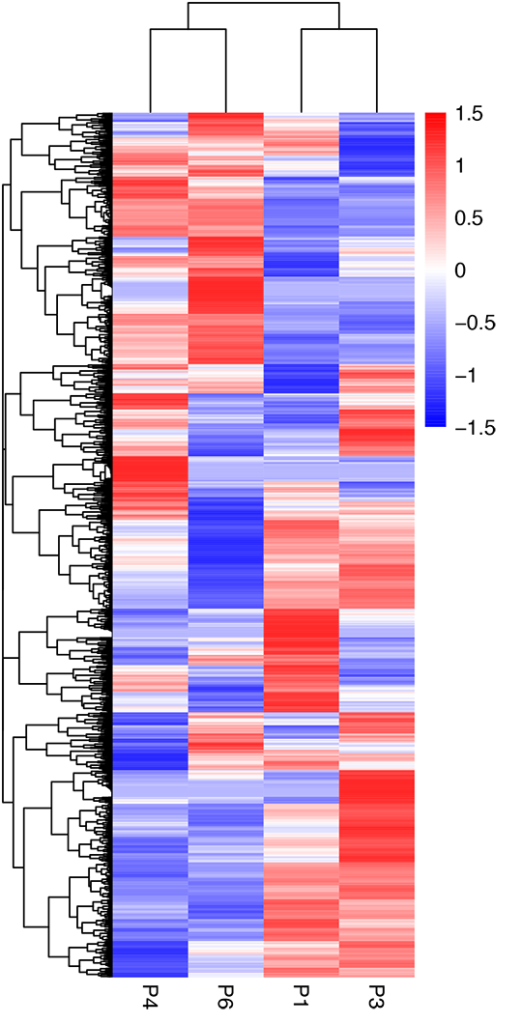
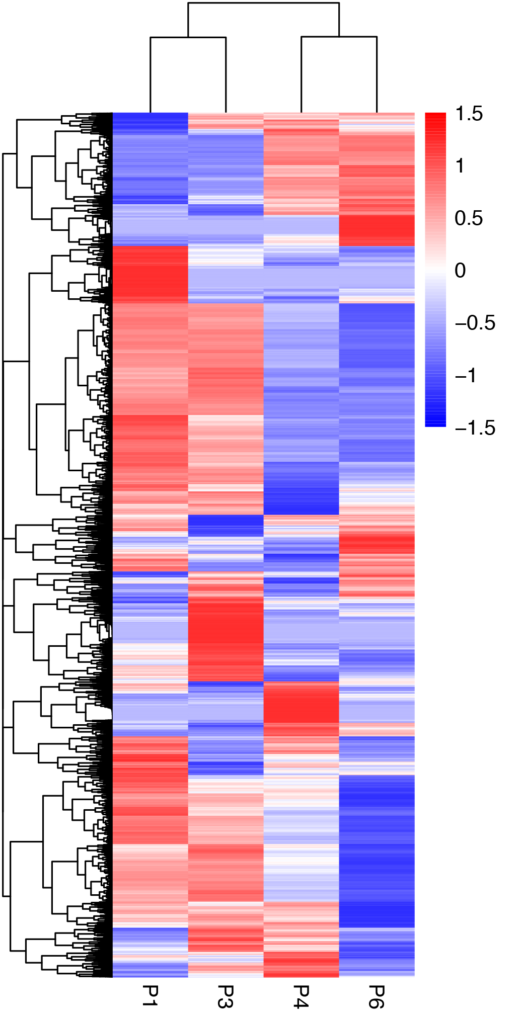
Clustering of mRNA
Clustering of lncRNA
Clustering of TUCP