Understanding the Advantages of Long Non-coding RNA Sequencing
Introduction to non-coding RNA
Non-coding RNAs (ncRNA) are a large class of RNA molecules that play a pivotal role in many biological processes, by being one of the three main contributors to epigenetic changes along with DNA methylation, and histone modification. Non-coding RNA contributes to these epigenetic changes through transcriptional regulation of both chromatin modification and post-transcriptional processing.1
Non-coding RNAs are comprised of many different kinds of RNA molecules. The most prevalent and diverse of which are long non-coding RNAs (lncRNA). They are typically over 200 nucleotides long and lack protein-coding potential.2 Long non-coding RNAs exhibit a huge range in size, shape, and function, and they are present within introns, antisense transcripts of protein-coding genes, enhancers, and other open genomic regions.3 Recent studies have unveiled a captivating array of advantages associated with studying lncRNA, including better understanding of the transcriptional landscape, chromatin remodeling, and epigenetic inheritance.2
Long non-coding RNA sequencing (lncRNA-Seq) is a powerful tool for studying lncRNAs’ functional role in diverse biological processes and human diseases. It enables the exploration of novel transcripts and discovery of regulatory mechanisms governing messenger RNA (mRNA) translation. In this blog, we will go deeper into the standard analysis process behind Novogene’s lncRNA sequencing and how it can be utilized in multiple applications.
1. LncRNA-Seq Solution at Novogene
Based on Illumina NovaSeq 6000 platform, Novogene’s lncRNA sequencing is designed for in-depth analysis of transcriptomes. It implements expert quality control at every stage of the sequencing process. Our lncRNA sequencing workflow is summarized as follows:
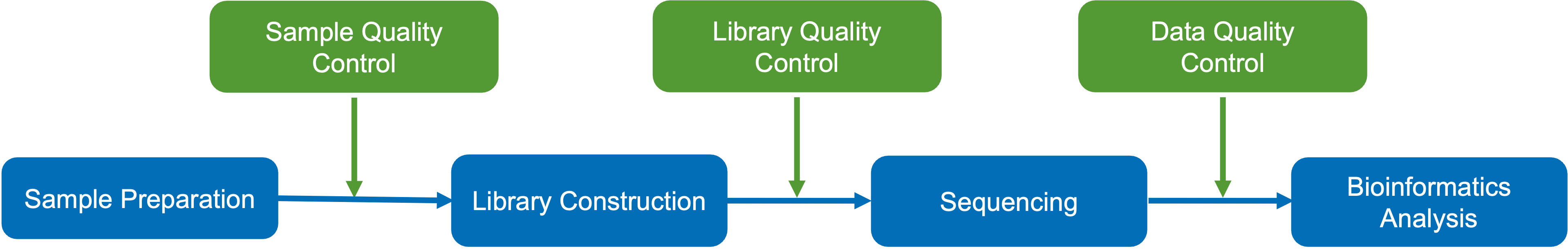
Figure 1. LncRNA sequencing workflow.
How are lncRNAs captured?
The lncRNA library construction workflow in Novogene is shown as follows. We provide a directional (strand-specific) library (250-300bp insert) with rRNA depletion and hence the lncRNA, mRNA and circRNA are all captured in the RNA sample.
Library preparation pipeline in Novogene:
- rRNA depletion
- Fragmentation
- Second strand cDNA synthesis
- End repair with a polyA-tail, and adapter ligation
- Enzyme digestion to remove the U-containing second strand of cDNA
- Size selection
- PCR enrichment
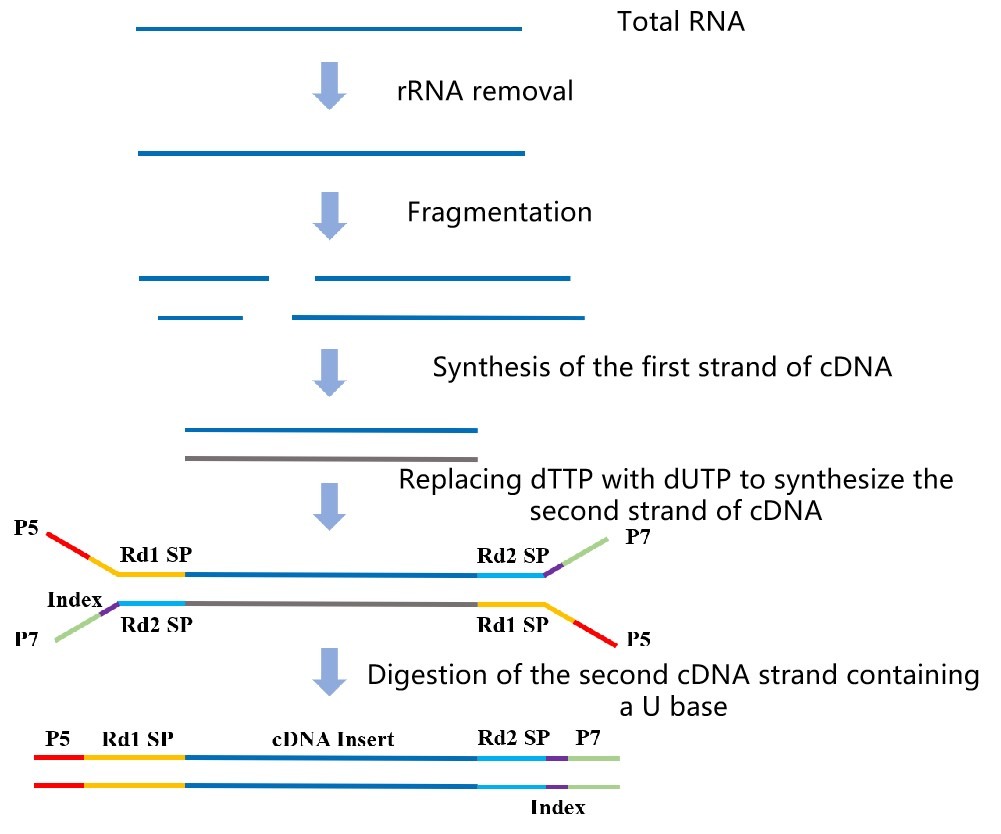
Figure 2. The lncRNA-seq library construction workflow.
2. Standard Analysis of Long Non-Coding RNA Sequencing
A routine lncRNA-seq project follows an established pipeline, which also includes mRNA in its analysis. The standard lncRNA sequencing pipeline of eukaryotic species (with a reference genome) is illustrated in Figure 3. The first step is ensuring that the raw data meet the quality control standards. The raw data are checked for error rate distribution, GC-content, and passed through data filtering.
The RNA molecules are then mapped to the reference genome, and structural variations can then be analyzed. Screening transcripts is essential for the next stage: quantification. Before that can occur, the screened transcripts are classified into two groups: known and novel transcripts. The lncRNA identification pipeline is shown in Figure 4.
After quantification, the final stage involves differential expression analysis of both mRNA and lncRNA. Target prediction of lncRNA can then also be used in functional enrichment analysis. Through enrichment analysis of differentially expressed genes, researchers can find out the associated biological functions or pathways are significantly associated with them, including GO Enrichment, KEGG Enrichment, etc.
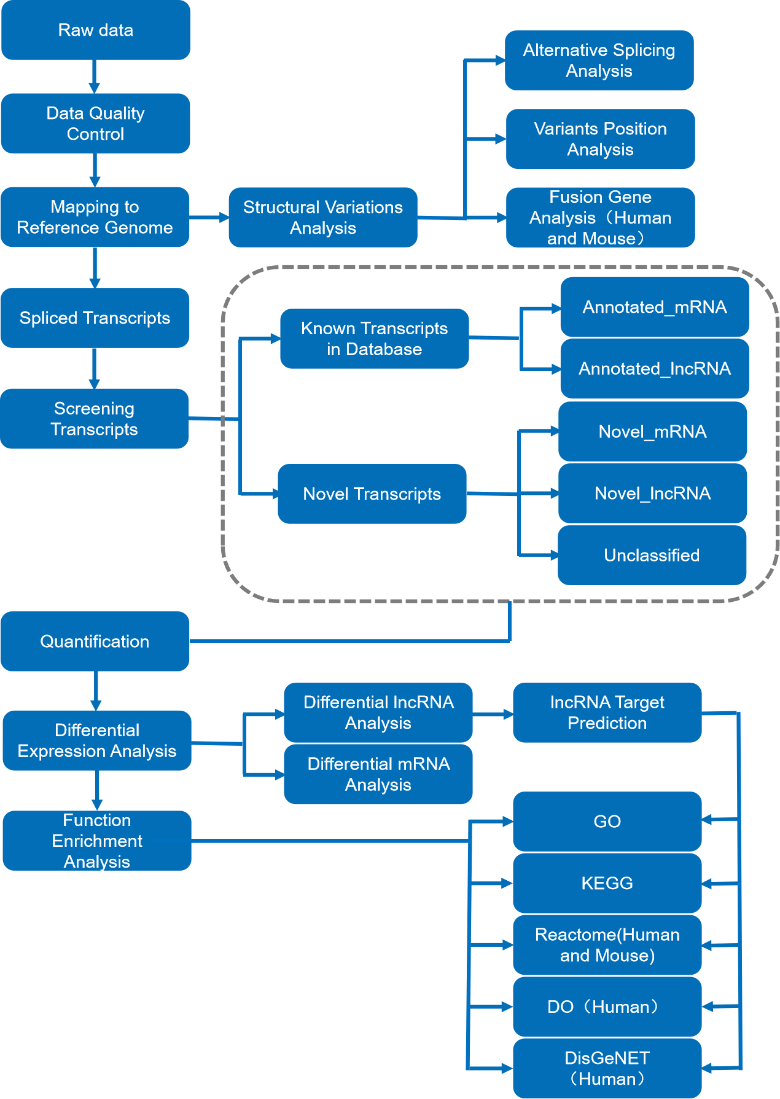
Figure 3. Long non-coding RNA sequencing analysis pipeline (mRNA included).
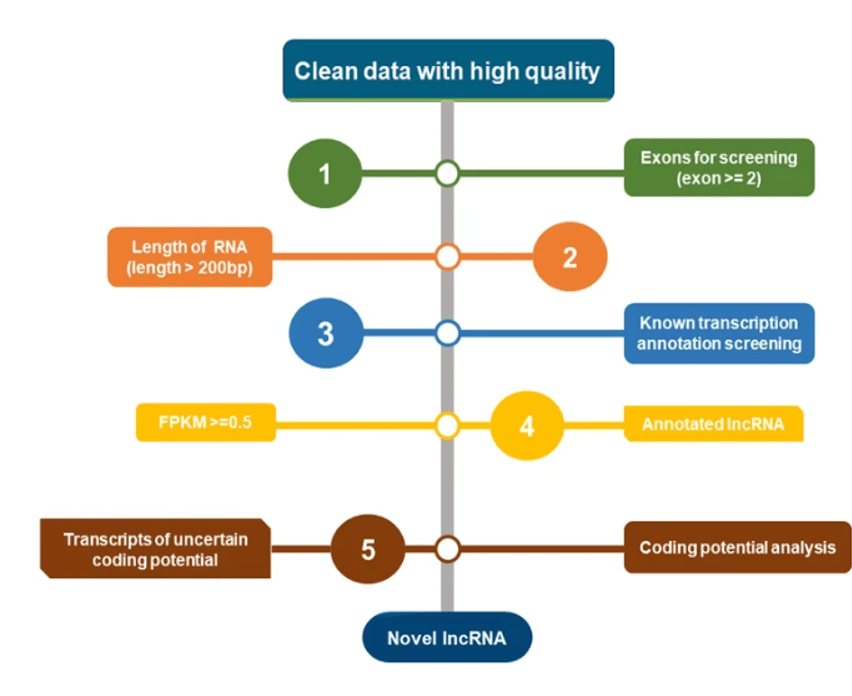
Figure 4. Long non-coding RNA identification pipeline.
3. Applications of Long Non-coding RNA Sequencing
There are various applications for lncRNA-seq, such as:
3.1 Identification of novel lncRNAs
- As mentioned before, after identifying transcripts that do not encode proteins as candidate lncRNAs, researchers can identify the specific features, including characteristic secondary structures, such as stem-loops and pseudoknots, or transcripts that are conserved across species.
- Validating novel ncRNAs can be done using a variety of experimental methods, such as Northern blotting or RT-PCR. Once a novel ncRNA is identified, its function can be studied using a variety of experimental methods, such as gene silencing or overexpression studies.
3.2 Analysis of differential expression of lncRNAs
- Statistical methods are used to calculate expression values for lncRNAs that are differentially expressed between the different groups or conditions. This is typically done using a software package such as DESeq2 or edgeR. Bear in mind that sequencing more samples in both control and experimental groups enhances the statistical power, providing a heightened level of confidence in identifying differentially regulated transcripts for achieving statistical significance. The differentially expressed genes can be visualized by a volcano plot (Figure 5). We also use the mainstream hierarchical clustering to cluster the fragments per kilobase of transcript per million reads (fpkm) values of genes, and homogenize the row (Z-score) (Figure 6).
- The results of the differential expression analysis can be validated using an independent method, such as quantitative real-time PCR (qPCR).
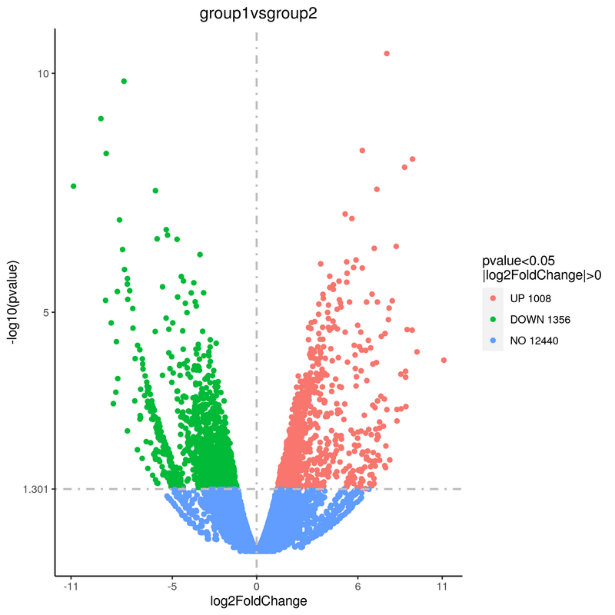
Figure 5. Differential Gene Volcano plot
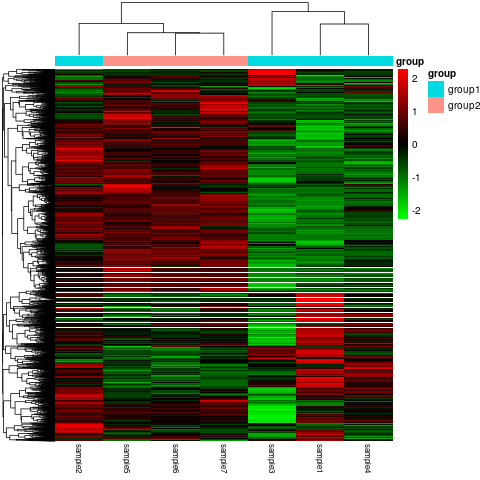
Figure 6. Differential expression gene clustering heatmap
3.3 Function enrichment analysis
- LncRNA target prediction: Target gene prediction can be performed for all lncRNAs through co-location and co-expression between lncRNA and protein-coding genes. Then, functional enrichment analysis (GO/KEGG) can be performed on target genes of different lncRNAs to predict the main functions of lncRNA (Figure 7-8).
- Differential lncRNA analysis: Differential lncRNAs can be displayed through the volcano plot and clustering heatmap, as shown above(Figure 5-6).
- Enrichment analysis of differential lncRNA target genes: To predict the function of lncRNA, GO and KEGG enrichment analysis can be performed for co-location and co-expression target genes of differential lncRNA, respectively.
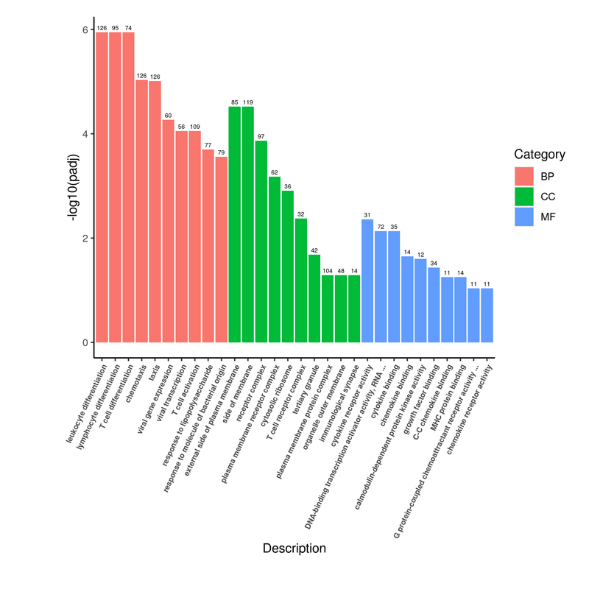
Figure 7. GO enrichment analysis histogram
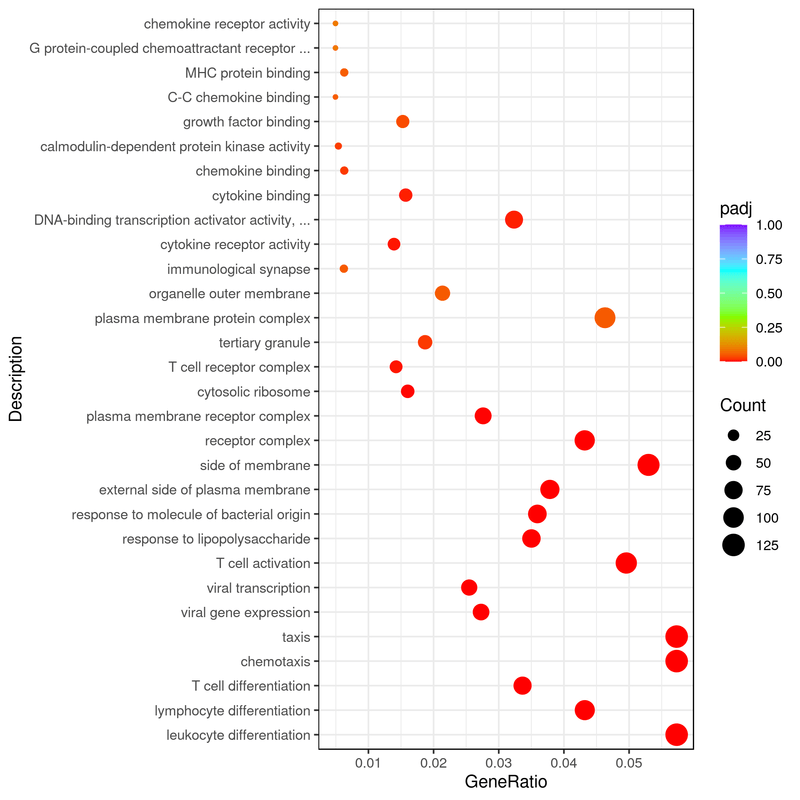
Figure 8. GO enrichment analysis scatter plot
4. Conclusion
Novogene provides comprehensive lncRNA-seq services characterized by high quality, affordable prices, and convenient solutions. Through lncRNA-seq analysis, the strand orientation and the regulatory association between lncRNA and targeted mRNA can be investigated, gaining unparalleled insights into lncRNA functions and interactions.
Whether you’re studying developmental biology, disease, or environmental stimuli, such as stress and infection, our lncRNA sequencing service is your key to uncover the potential of lncRNAs. So, get ready and push the boundaries of what’s possible with Novogene’s lncRNA-seq services!
References:
1. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009 Feb 20;136(4):629-41. doi: 10.1016/j.cell.2009.02.006. PMID: 19239885.
2. Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016; 937: 3–17. https://doi.org/10.1007/978-3-319-42059-2_1 PMID: 27573892
3. Li W, Huo Y, Ren Y, Han C, Li S, Wang K, He M, Chen Y, Wang Y, Xu L, Guo Y, Si Y, Gao Y, Xu J, Wang X, Ma Y, Yu J, Wang F. Deciphering the Functional Long Non-Coding RNAs Derived from MicroRNA Loci. Adv Sci (Weinh). 2023 Nov;10(33):e2203987. doi: 10.1002/advs.202203987. Epub 2023 Oct 17. PMID: 37849233; PMCID: PMC10667839
4. Gapp, K., van Steenwyk, G., Germain, P.L. et al. Alterations in sperm long RNA contribute to the epigenetic inheritance of the effects of postnatal trauma. Mol Psychiatry 25, 2162–2174 (2020). https://doi.org/10.1038/s41380-018-0271-6
