Introduction to Enzymatic Methyl Sequencing
DNA methylation at the C5 position of cytosine plays a crucial role in gene expression and chromatin remodelling. As one of the most important research areas in epigenetics, methyl-seq tech is .With the development of high-throughput sequencing technology, WGBS has gradually become the gold standard for DNA methylation sequencing.
Enzymatic Methyl Sequencing has been developed to overcome the shortcomings of WGBS, an enzyme-based method for the detection of 5mC and 5hmC at the single base level. The enzymatic conversion method is gentler and causes less DNA damage, thereby avoiding DNA molecule cleavage and loss.
Enzymatic methylation sequencing Highlights
- Enzyme based method Minimizes damage to DNA molecules
- Less DNA input required
- Less sequencing depth required
- Superior performance compared to bisulfide sequencing
- cfDNA is acceptable
Enzymatic methylation sequencing Specifications: DNA Sample Requirements
| Sample Type | Amount | Purity or fragment size |
| Genomic DNA | ≥ 50 ng | Fragments are above 5000 bp, and mainly above 13000 bp, no degradation, no contamination, no EDTA |
| cfDNA | ≥ 50 ng | Agilent 2100 peak at 170bp and integer multiples, no genomic contamination, no contamination, no EDTA |
Enzymatic methylation sequencing Specifications: Sequencing and Analysis
| Platform Type | Illumina NovaSeq platform |
| Read Length | Paired-end 150 bp |
| Sequencing Depth | ≥ 20× coverage for the species with reference genome |
| Standard Data Analysis |
Data Quality Control mCs detection, methylation level calculation Methylation level and frequency distribution Differentially Methylated Site (DMS) detection Differentially Methylated Regions (DMRs), Differentially Methylated Promoter (DMPs) detection and annotation Function enrichment of DMR-associated genes and DMP-associated genes Visualization of BS seq data Comparative analysis (among samples) |
| Co-Analysis with mRNA-seq (Integrated Analysis of Methylome and Transcriptome) |
The distribution of gene methylation level and expression level at the chromosome level, and the distribution of methylation level and expression level at the upstream and downstream levels of genes. The relationship between DMR related gene’s expression and methylation modification The intersection of DMR related genes and differentially expressed genes, and display the methylation level, expression amount and annotation information of the intersection genes in different forms. Functional analysis of DMR related genes and differentially expressed genes, including GO, KEGG, and Motif Analysis. |
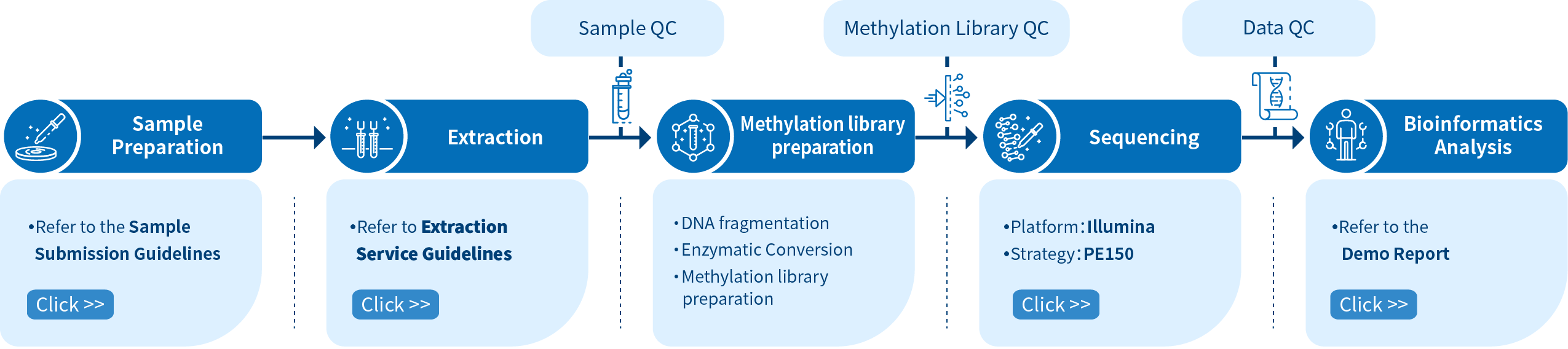
Project Workflow of Novogene enzymatic methylation sequencing Service
The Novogene enzymatic methylation sequencing service comprises four steps: Sample preparation, library preparation, sequencing, and bioinformatics analysis, DNA extraction service is also available. The enzymatic methylation sequencing conversion reaction is a two-step process: 1) TET2 and T4-BGT Protection of 5mC and 5hmC. 2) APOBEC then deaminates cytosines into uracils. Please contact us for more information about your enzymatic methylation sequencing projects.
To guarantee the accuracy and reliability of sequencing data, Novogene audits every experimental step through quality control, fundamentally ensuring high-quality data output from sampling to the final data report. This commitment to high-quality data is essential for the correctness, comprehensiveness, and credibility of bioinformatics analysis.
-
A genomic and epigenomic atlas of prostate cancer in Asian populations
NatureIssue Date: 25 March 2020IF: 42.778DOI: 10.1038/s41586-020-2135-x
-
Plant Biotechnology JournalIssue Date: 10 August 2017IF: 8.154DOI: 10.1111/pbi.12820
-
BioinformaticsIssue Date: 27 January 2017IF: 5.610DOI: 10.1093/bioinformatics/btx040
-
The Plant CellIssue Date: 24 August 2016
-
Genome BiologyIssue Date: 04 March 2014IF: 10.806DOI: 10.1186/gb-2014-15-3-r49
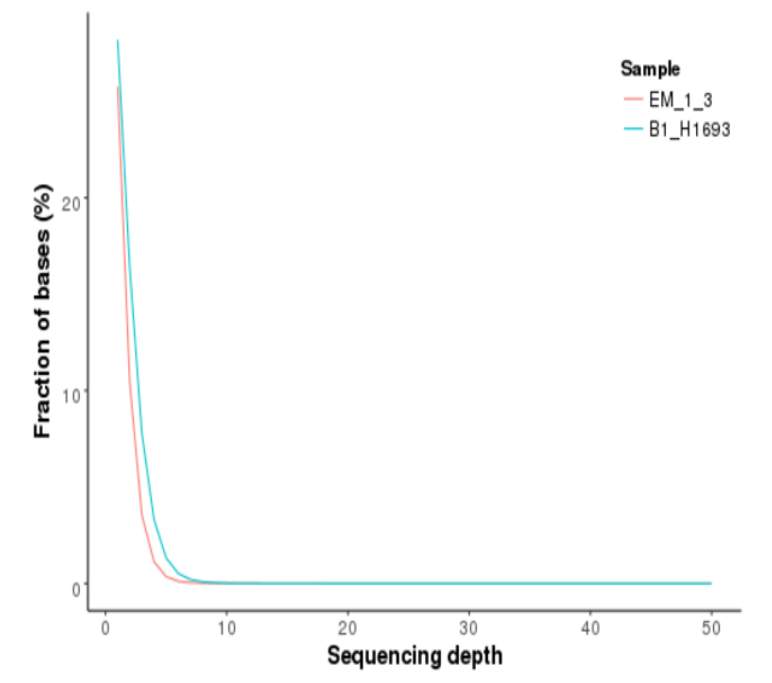
Distribution of Genome Coverage
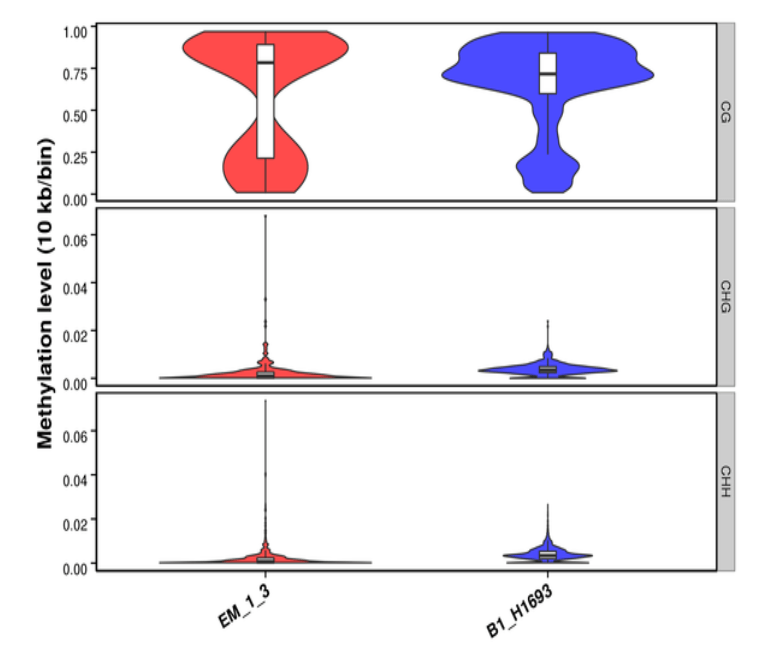
Methylation Level Distribution on Whole Genome
Heatmap Analysis for Methylation Levels of Gene Functional Region
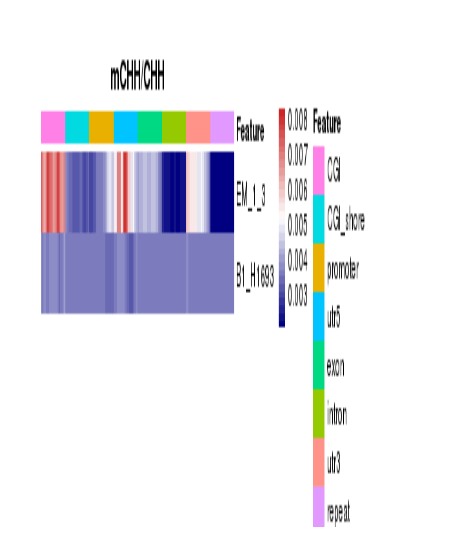
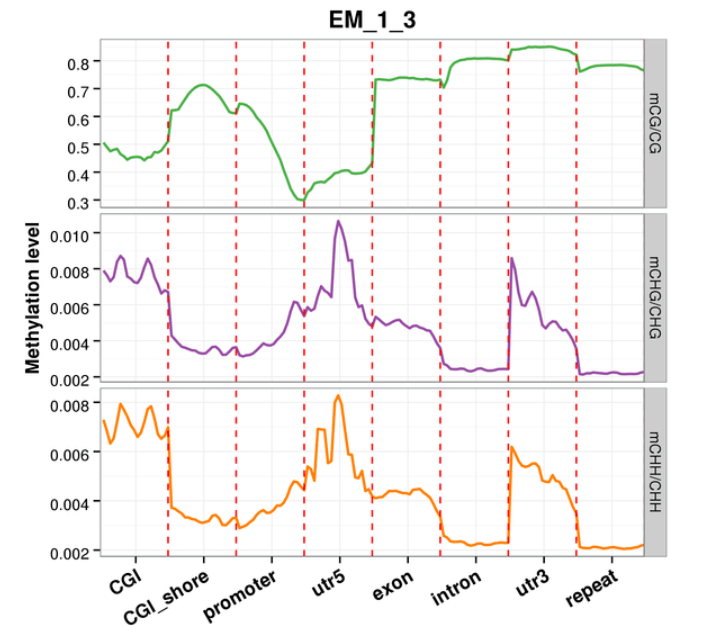
Methylation Level Distribution at Functional Genetic Elements
Circos Plot for DMR Condition in three contexts (CG, CHG, CHH)
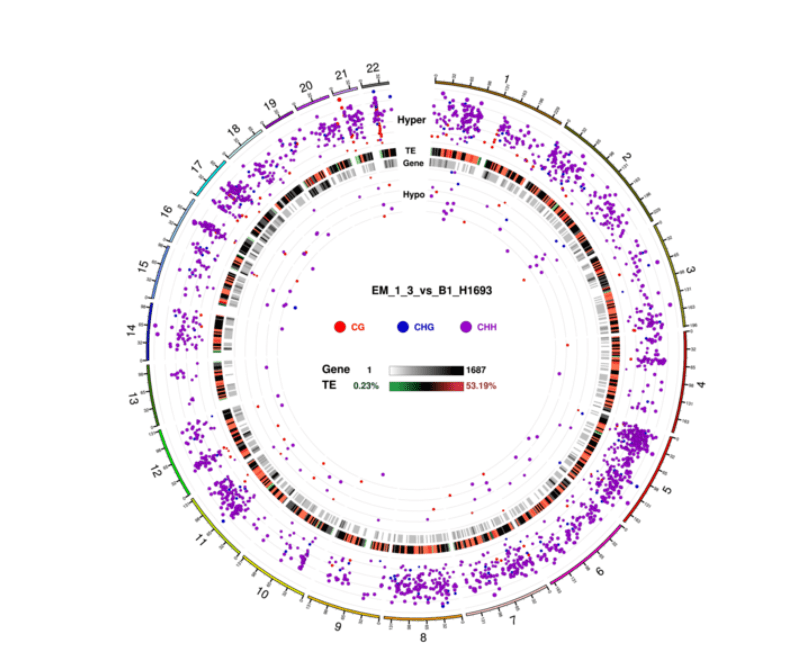
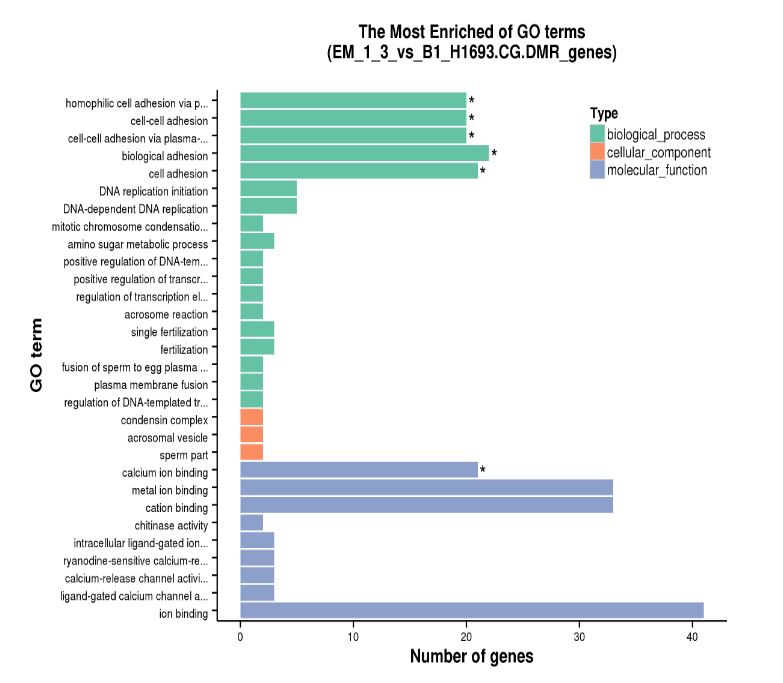
Diagram of GO Enrichment