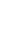
Comprehensive genomic profiling for 484 cancer-related genes to help you give the right treatment for your patients
The Novogene Precision Medicine 2.0 (NovoPM™ 2.0) comprehensive genomic profiling test for solid tumors is a hyphenate sequencing (NGS)-based assay that analyzes 484 genes for clinically important alterations at the DNA level. These genes are known to be relevant for the diagnosis and/or treatment of various solid tumors according to National Comprehensive Cancer Network (NCCN) Guidelines and the medical literature. All tests are performed in our CAP-accredited lab and analyzed by our in-house bioinformatics specialists. Report generated provides tailored treatment recommendations for your patients.
NovoPM™ 2.0 is developed based on NovoPM™ 1.0 with updated gene list and enhanced analytical performance.
Parameters
| Cancer type | All Solid Tumors |
| Sample type | Extracted DNA FFPE Whole blood Blood Pasma Extracted cfDNA |
| Turnaround time | ≤ 2 weeks |
Novogene Quality Assurance
Comprehensive Results
NovoPM™ 2.0 interrogates the complete coding regions of 468 genes and the introns of 43 genes. It can detect all four types of genomic abnormalities(SNV, InDel, CNV and Fusion) and also generate results (TMB, bTMB and MSI)that can help guide cancer immunotherapies.
Reliable Performance
Comprehensive analytical validation has been completed and the test includes rigorous quality control measures.
Ultra-fast Sample Importation (when necessary)
As a high-tech life science company in Beijing, Novogene can utilize a government-authorized "green channel" for ultra-fast importation of clinical samples through the Beijing Customs, when necessary.
Competitive Price
With one of the world's largest sequencing capacity and superior process efficiency, Novogene offers highly competitive prices for all services.
Who Benefits from NovoPM™ 2.0 ?
- Individuals with advanced or late stage cancers.
- Individuals who have tested negative for single gene mutations and require a more comprehensive tumor profiling.
- Individuals with cancers of unknown primary origin.
- Individuals with inadequate amount of tissue or unobtainable tissue for biopsy.
- Individuals who may prefer non-invasive testing or are not suitable for invasive surgical procedure.
Gene Targets
NovoPM™ 2.0 interrogates the complete coding regions of 468 genes and the introns of 43 genes for all four types of genomic abnormalities: SNV, InDel, CNV and fusion. A comparison of the gene lists between NovoPM™ 2.0 and the FDA-approved FoundationOne CDx is shown in Figure 1. The mutation status of some of these genes can guide the potential application of multiple FDA-approved targeted and/or immunotherapies as shown in Table below. The other genes are also analyzed for their relevance to the diagnosis and/or treatment of various solid tumors according to the medical literature. The clinical interpretation of detected mutations in those genes is done according to Novogene’s comprehensive in-house oncology knowledgebase constructed based on public resources including GeneCards, CKB, OncoKB, ClinVar PMC, Drugs@FDA, Drug Information Portal (NIH), Selleck, PharmGKB, DGIdb, DRUGBANK, Drugs.com, ClinicalTrials.gov, ICTRP, ChiCTR, KEGG and Cell Signaling. In addition, our oncology knowledgebase incorporates ~1,000 germline BRCA1/2 variants previously reported to be unique in Chinese breast cancer and ovarian cancer patients1.

Figure 1. NovoPMTM 2.0 vs. FoundationOne CDx
Table 1. Gene targets in NovoPMTM 2.0 that are associated with FDA-approved or NCCN-recommended therapies
| Indication | Biomarkers | FDA-approved or NCCN-recommended therapies |
|
Non-Small Cell Lung Cancer
|
EGFR 19Del and L858R | Gilotrif® (afatinib), Iressa® (gefitinib) or Tarceva® (erlotinib) |
| EGFR T90M | Tegrisso® (osimertinib) | |
| ALK rearrangements | Alecensa® (alectinib), Xalkori® (crizotinib), Zykadia® (ceritinib), Alunbrig® (brigatinib) or Lorbrena® (Lorlatinib) | |
| BRAF V600E | Tafinlar® (dabrafenib) + Mekinist® (trametinib) | |
| ROS1 fusion | Xalkori® (crizotinib) or Rozlytrek® (entrectinib) | |
| TMB and bTMB | Opdivo® (nivolumab), Keytruda® (pembrolizumab), or Tecentriq® (atezolizumab) | |
| Melanoma | BRAF V600E or V600k | Tafinlar® (dabrafenib), Tafinlar® (dabrafenib) + Mekinist® (trametinib), Zelboraf® (vemurafenib) or Braftovi® (encorafenib) + Mektovi® (binimetinib) |
|
Breast Cancer
|
ERBB2 (HER2) amplification | Herceptin® (trastuzumab), Perjeta® (pertuzumab), Tykerb® (lapatinib) or Nerlynx (neratinib) |
| gBRCA alterations and ERBB2 (HER2) negative | Talzenna® (talazoparib) or Lynparza® (olaparib) | |
| PIK3CA alterations | Piqray® (Alpelisib) | |
|
Colorectal Cancer
|
RAS wild-type | Erbitux® (cetuximab), Vectibix® (panitumumab) or Stivarga® (regorafenib) |
| MSI-H | Keytruda® (pembrolizumab), Opdivo® (nivolumab) or Opdivo® (nivolumab) + Yervoy® (ipilimumab) | |
| Ovarian Cancer | BRCA1 and BRCA2 alterations | Lynparza® (olaparib) or Rubraca® (rucaparib) |
|
Solid Tumors
|
MSI-H and TMB | Keytruda® (pembrolizumab) |
| NTRK1/2/3 fusions | Vitrakvi® (larotrectinib) or Rozlytrek® (entrectinib) |
Sample Requirements
| Sample Type | Sample Requirement |
| FFPE | Approximately ten 4-µm sections, each with tissue area ≥ 25 mm2 and tumor content ≥ 20%. |
| Whole blood | 10 mL (for ctDNA and white blood cell DNA extraction) |
| Blood Plasma | 4 mL (for ctDNA extraction) |
Testing Workflow