Introduction to Single-Cell Technology: Fundamentals and Historical Development

Single-cell RNA sequencing (scRNA-seq) first emerged in 2009, marking the beginning of the single-cell omics era. In recent years, it has become a groundbreaking tool in biological research, enabling scientists to explore the intricate heterogeneity of individual cells—something traditional bulk sequencing could not achieve. Single-cell technologies now span multiple omics levels, including genome sequencing, DNA methylation profiling, and ATAC-seq for chromatin accessibility. Among these, single-cell transcriptome sequencing (scRNA-seq) stands out as the most prominent technique. Its rise has not only overcome longstanding technical challenges in biology but has also gained rapid popularity in the research community due to its cost-effectiveness and transformative insights. Today, single-cell sequencing continues to drive a new wave of discovery in life sciences, offering unprecedented view and understanding of cellular complexity.

Figure 1: Current Major Single-Cell Sequencing Platforms
I. The Purpose of Single-Cell Sequencing
Single-cell sequencing technology, with its exceptional resolution and depth, overcomes the limitations of traditional sequencing methods in analyzing tissue compartmentalization and cellular heterogeneity. In many research scenarios, the focus is on small regions within a tissue or specific cell populations—details that conventional transcriptome sequencing often struggles to capture accurately. Traditional approaches typically rely on analyzing bulk samples extracted from an organ, which can underrepresent the target tissue and obscure critical biological information.
In fact, tissue complexity arises largely from cellular diversity. While conventional transcriptomics provides an overall gene expression profile, it cannot easily distinguish subtle differences between distinct cell types or states. This limitation is especially pronounced when studying rare or specialized cells.
Single-cell sequencing, however, addresses these challenges by profiling individual cells, uncovering nuanced differences, and offering a high-resolution view of cellular heterogeneity. For researchers seeking a deep, precise understanding of cellular behavior and diversity, single-cell sequencing is an indispensable tool that enables insights previously unattainable with bulk approaches.
II. General Principles of Single-Cell Sequencing Technology
Molecular Quantification
Current methods for RNA quantification in single-cell sequencing are primarily based on two approaches: full-length and tag-based. Full-length methods capture complete RNA transcripts, providing uniform coverage across the entire molecule and allowing detailed analysis of isoforms and splice variants. Tag-based methods, in contrast, sequence only the 5′ or 3′ ends of RNA using specialized probes, which enables efficient quantification of transcript abundance across large numbers of cells.
Cell Capture
The most widely used approaches for isolating individual cells are based on microwells, microfluidics, and droplet-based platforms. Microwell systems physically separate cells into small wells for downstream processing. Microfluidics platforms guide cells through narrow channels to capture and process them individually. Droplet-based methods encapsulate single cells in nanoliter droplets, allowing high-throughput analysis with minimized cross-contamination. Each method balances throughput, cost, and sensitivity, providing flexibility depending on the experimental needs.
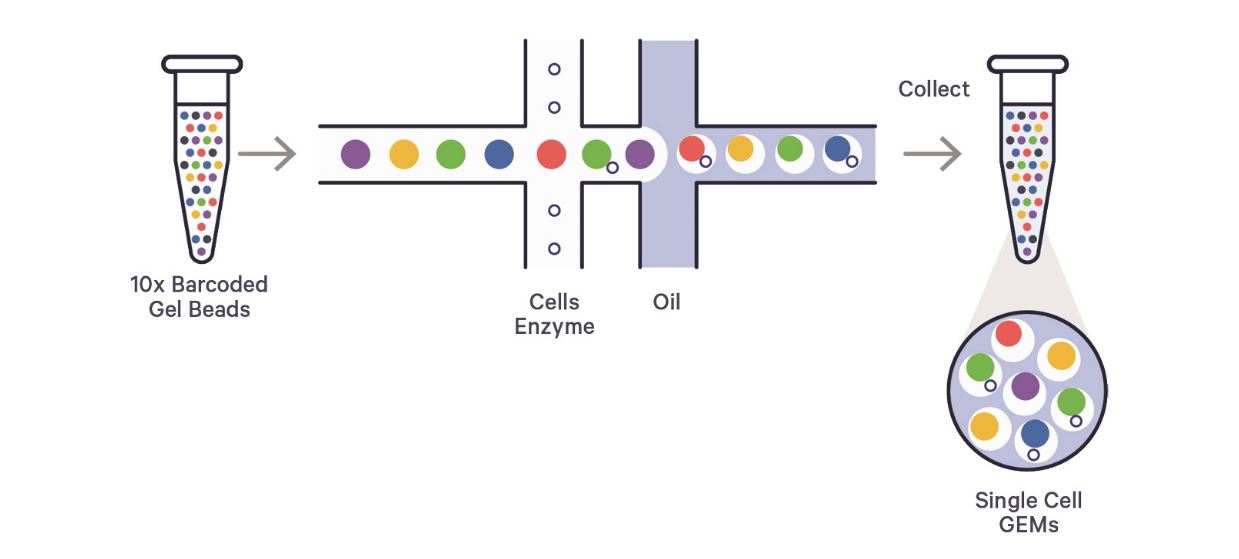
Figure 2: Schematic Diagram of Cell Capture Method Based on Microfluidics
III. 10x Genomics: A Leading Single-Cell Sequencing Platform
10x Genomics, known for its high performance and versatile applications, is currently the most widely used single-cell sequencing platform in academic research. To illustrate the single-cell sequencing workflow, we will use 10x Genomics 3′ transcriptome sequencing as an example to provide a brief overview of the entire process.
This technology cleverly uses droplets as isolation units, encapsulating individual cells within water-in-oil microdroplets. Building on the Drop-seq principle, 10x Genomics establishes an efficient cell isolation system within microfluidic channels. In this setup, each cell is separated individually, ensuring the specificity and accuracy of downstream reactions.
The figure below illustrates the key steps of 10x scRNA-seq, including:
- Cell suspension preparation
- Library construction
- Sequencing
- Data analysis
- Visualization
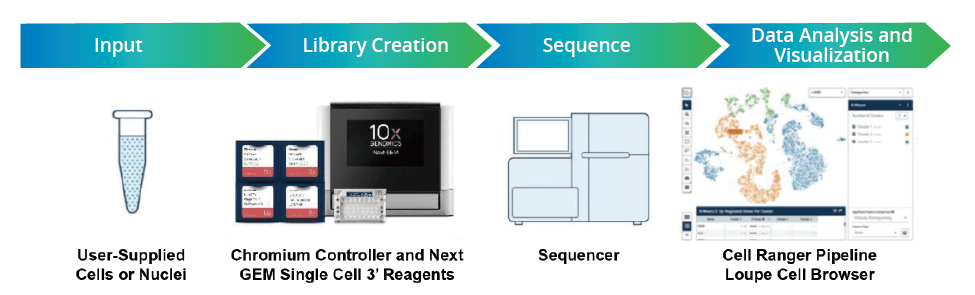
Figure 3: 10x Genomics scRNA-seq Workflow
The library preparation process can be divided into four steps:
A. Bead Preparation:
Specialized beads provided by 10x Genomics have key molecules attached to their surface for subsequent reverse transcription, including adapters, barcodes, Unique Molecular Identifiers (UMIs), and other sequences. These beads form the foundation for building single-cell libraries, providing the necessary molecular tools for subsequent operations.
B. Bead and Cell Encapsulation:
Beads flow through the aqueous channel within a microfluidic chip, passing through two critical "crossroads." At the first intersection, beads encounter a channel carrying cells and reverse transcription amplification reagents; some beads adsorb cells while the liquid environment becomes enriched with the required enzymes. At the second intersection, an incoming oil phase mixes with the aqueous phase, forming a water-in-oil emulsion. Due to the hydrophilic nature of the beads, they, along with the adsorbed cells, become encapsulated within water droplets, forming structures known as GEMs (Gel Bead-in-Emulsions).
C. Intracellular Reactions:
Within the GEM microenvironment, cells are lysed, RNA is released, reverse transcription occurs, and cDNA is amplified. This step is completed within the closed microdroplet, ensuring the independence and accuracy of molecular information from each cell.
D. cDNA Amplification and Sequencing Preparation:
After amplification, the cDNA from each cell carries unique molecular identifiers and is prepared for high-throughput sequencing. These cDNA molecules, bearing the cell’s gene expression information, are sent to the sequencing platform to reveal the cell’s genetic blueprint.
The careful design and execution of these steps ensure that the 10x Genomics platform delivers high efficiency and accuracy, offering researchers a powerful tool to investigate cellular heterogeneity in depth.
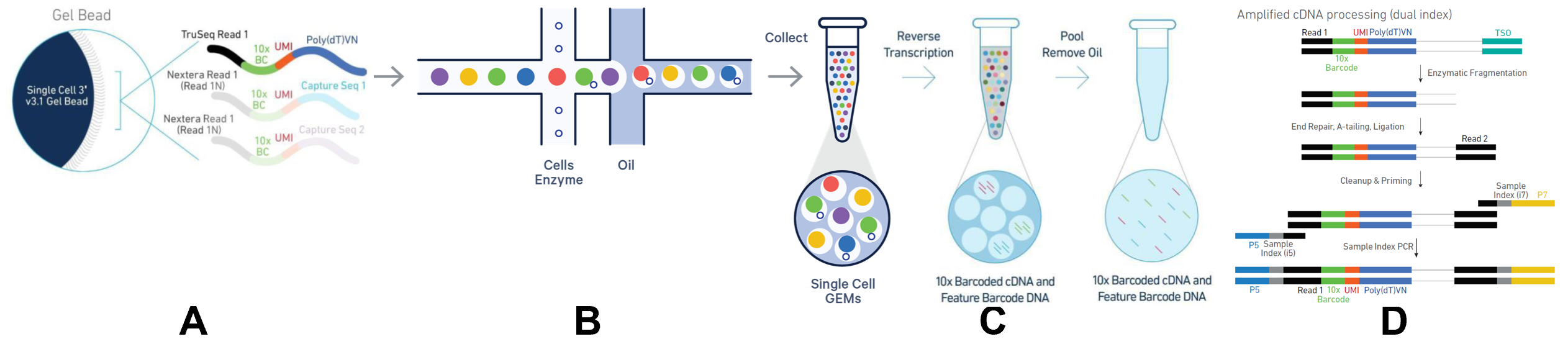
Figure 4: Schematic Diagram of 10x Genomics scRNA-seq Library Preparation Principle
Following sequencing, complex cellular information can be deeply mined and interpreted using bioinformatics approaches.
A. Standard Analysis Methods
- Clustering: Groups cells based on their gene expression patterns using computational algorithms, revealing similarities and differences among cells.
- Differential Gene Expression Analysis: Identifies genes with significantly different expression levels across cell populations or states, offering insights into cell function and identity.
- Enrichment Analysis: Examines the overrepresentation of specific biological processes or pathways within cell populations to better understand their functional characteristics.
B. Advanced or Custom Analysis Features
- Subclustering: Further refines broader cell populations to identify smaller subpopulations with distinct functions.
- Pseudotime Analysis: Models dynamic changes in cell states, revealing temporal patterns during development or response processes.
- Receptor-Ligand Analysis: Investigates interactions between cell surface receptors and ligands, providing insights into mechanisms of cell-cell communication.
By combining single-cell sequencing with in-depth bioinformatics analyses, researchers can uncover not only the intrinsic properties of individual cells but also their interactions and dynamic changes over time. The versatility and customizability of these analytical methods are among the technology’s most compelling features, providing unprecedented depth and breadth for exploring complex biological systems.
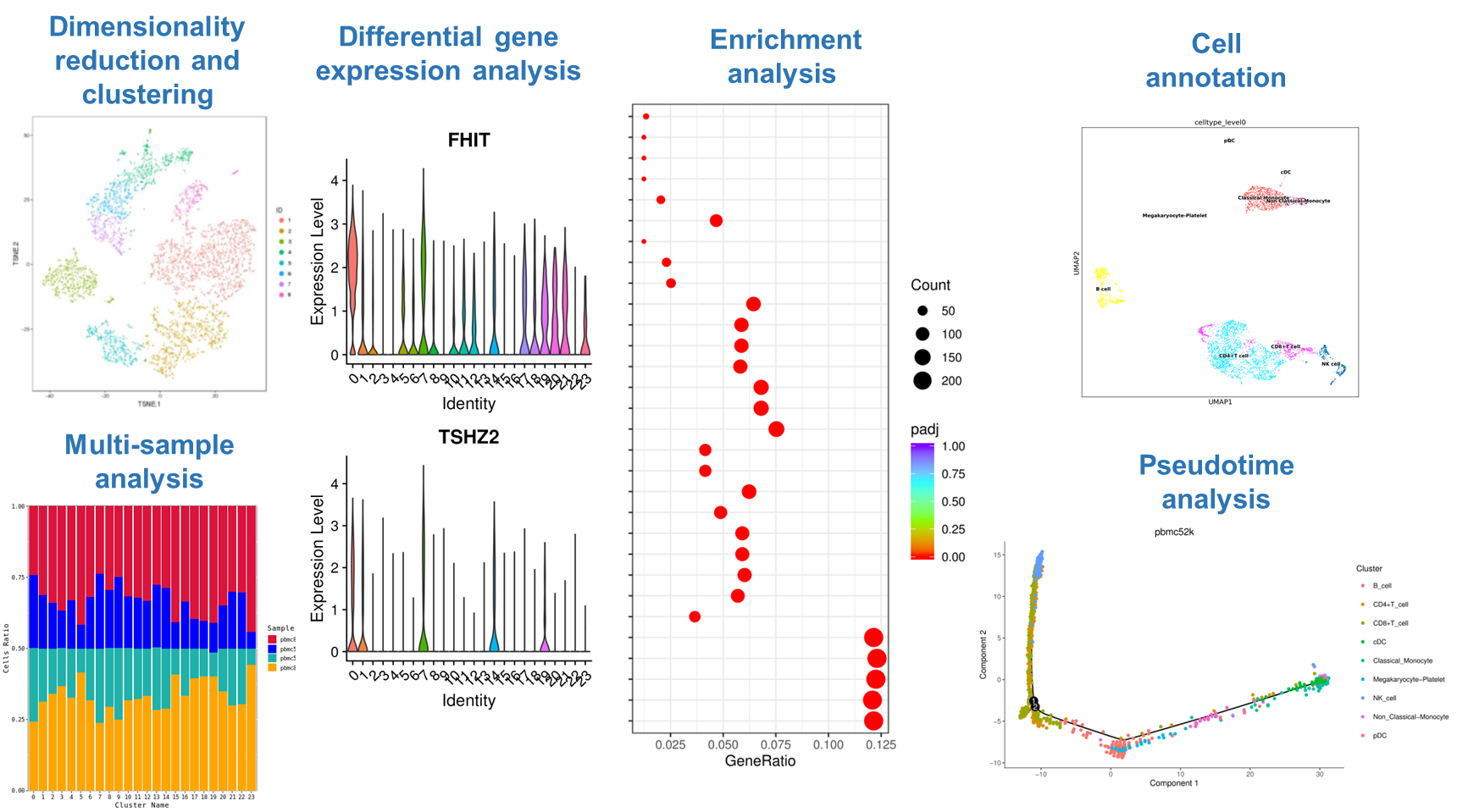
Figure 5: Example Visualization of Single-Cell 3′ Transcriptome Sequencing Bioinformatics Analysis
Single-cell sequencing technology, with its high resolution, offers researchers a powerful tool to explore biological processes at the cellular level. It not only deepens our understanding of cellular diversity but also enables the discovery of subtle molecular patterns, driving scientific breakthroughs and opening new avenues for disease treatment and health maintenance.
By providing detailed, cell-specific gene expression information, single-cell sequencing overcomes the limitations of bulk transcriptome analysis. Its unique strengths include revealing cellular heterogeneity, dissecting cell differentiation and developmental pathways, identifying novel cell types, precisely analyzing disease mechanisms, and studying cell-cell communication. Already, single-cell technology has made a transformative impact across diverse fields, including biology, medicine, and agriculture.
Why Choose Novogene for Single-cell RNA Sequencing (single-cell RNA-Seq or scRNA-Seq)?
- Proven Expertise: With over 200,000 successfully sequenced samples, Novogene delivers great project results at industry-leading turnaround times. We excel at handling challenging sample types, including nerve and adipose cells.
- Enhanced Sample Processing: We offer a diverse range of sample processing capabilities, including nuclei extraction and specialized pipelines for frozen tissues. This ensures high-quality gene expression data in Single-cell RNA Sequencing (single-cell RNA-Seq or scRNA-Seq) projects.
- Certified Excellence: As a 10x Genomics Certified Service Provider, we leverage the advanced Chromium X platform combined with GEM-X technology for superior reproducibility and efficiency.
- Cost-Effective Solutions: We have state-of-the-art high throughput sequencing platforms, coupled with expert support, ensure exceptional data quality and provide cost-effective solutions for single-cell projects.

