Four Powered Online Tools for Genomic Analysis and Visualization (IV) – circos
IV.circos – circos diagram
Circos is a command-line software that can be used for visualizing data and information. The online application is suitable for creating rigorously informative infographics with texture and aesthetic appeal. It can create both low-resolution bitmaps for web-page integration, as well as high-resolution publication standard images.
Circos specifically visualize data in a circular layout which makes it the perfect tool for exploring and representing relationships between objects and positions. With its high data-to-ink ratio, circos creates images that are highly attractive. Each element in the image is modular and controllable, and allows the user to tailor the focus points and details catered to the audience’s appeal. Get the figure from http://mkweb.bcgsc.ca/tableviewer/visualize/.
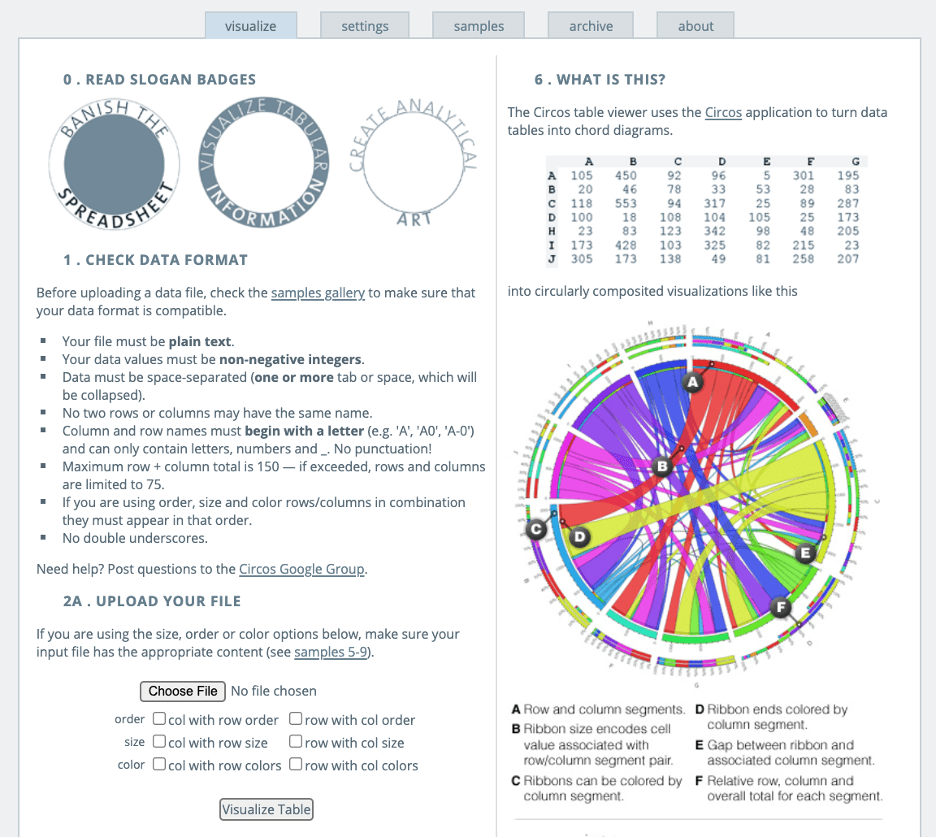
For input, circos takes GFF-style data files and Apache-like configuration files both of which can be easily generated by automated tools. The configuration is modular, and parameter blocks are reusable when imported from multiple files. The output images hence generated can be created in PNG (8 or 24 bit) or SVG formats. Intuitive and easy to play around, circos definitely is a tool that makes a biological scientist’s work more palatable and attractive for a wider audience[1].
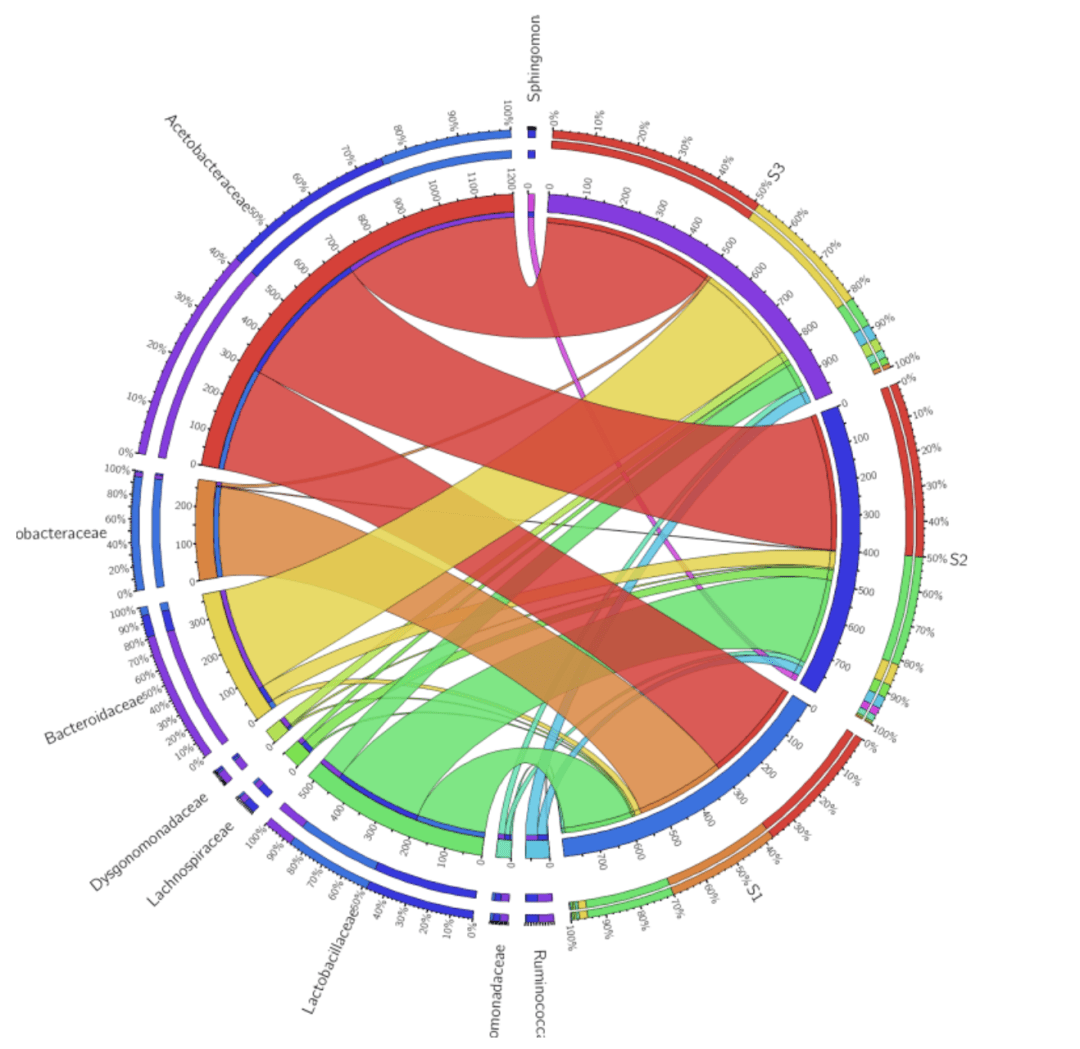
Taking the analysis of the microbial abundance and species diversity as an example, here shows demos to explain the function of drawing the circos diagram.
1) Data Preparation
The data used are the relative abundance of species in each sample, as follows (S1, S2, S3 are sample names in columns, and Acetobacteraceae, and other species classifications are in rows). Please note that the data formats are required in a non-negative integer.
| OTU | S1 | S2 | S3 |
| Acetobacteraceae | 300 | 400 | 450 |
| Sphingomonadaceae | 0 | 18 | 0 |
| Bacteroidaceae | 19 | 50 | 300 |
| Ruminococcaceae | 3 | 27 | 35 |
| Pseudomonadaceae | 7 | 15 | 19 |
| Lactobacillaceae | 200 | 250 | 70 |
| Lachnospiraceae | 5 | 30 | 15 |
| Dysgonomonadaceae | 5 | 5 | 29 |
| Arcobacteraceae | 260 | 3 | 10 |
2) Diagram Generation & Saving
Upload your data file and click button of “Visualize Table”. Then you will get the Circos image ready. By setting the segments, labels and other parameters, the layout of the images will be optimized based on your own needs.
Reference
[1] Krzywinski, Martin et al. “Circos: an information aesthetic for comparative genomics.” Genome research vol. 19,9 (2009): 1639-45. doi:10.1101/gr.092759.109
