Four Powered Online Tools for Genomic Analysis and Visualization (III) – iTOL
III. iTOL – Evolutionary Tree Beautification
In biology and related fields of science, phylogenetic trees are vital tools for contextualization and representation of various data types. iTOL (abbr: interaction Tree Of Life) is an interactive tool that integrates online display, annotation and management of evolutionary trees. iTOL is an online tool that can be accessed with any modern web browser, and it is implemented in pure Javascript. This tool enables users to beautify the phylogenetic tree they aim to construct. As they draw, they can freely adjust the color, shape and font of the branches and labels. iTOL can display different datasets simultaneously and offer customized control of position, size and color according to individual needs. The output can finally be exported as high-quality bitmaps and vector graphics.
iTOL supports different common formats of phylogenetic trees: Newick, Nexus and phyloXML. Phylogenetic placement files and annotation files alike can be uploaded directly and annotated for taxonomic, frequency and sequence alignment visualizations. Any extra data that is relevant can be provided in plain text files, and by simple drag-and-drop mechanism, visualized in the user’s web browser. On top of standard display formats (rectangular, circular and unrooted) the latest version of iTOL supports slanted phylogram display mode as well. The trees such built may be manipulated in several ways, and it is possible to interactively edit by deleting or moving single nodes or whole clades. The clades can be pruned, collapsed and various parameters like branch length distance be adjusted either manually or automatically. Since raw FASTA multiple sequence alignments are also supported by the tool, consensus sequences and residue conservation graphs also can be easily calculated and presented.
Hence, allowing for visualization trees with even more than 50,00 leaves, iTOL is a powerful tool at a biologist’s disposal for beautifying the vast genomic data[1]. Get access to use at: http://itol.embl.de/index.shtml.
Step 1. Raw File Preparation
Files formats with plain texts only, such as Newick, Nexus, PhyloXML, Text, and Jplace, can be recognized by the tool.
Step 2. Annotation Dataset
Download the annotation templates of evolutionary tree from http://itol.embl.de/help.cgi#annot and prepare datasets accordingly.
Step 3. File Upload
Upload the original phylogenetic tree file to the “Tree file”.
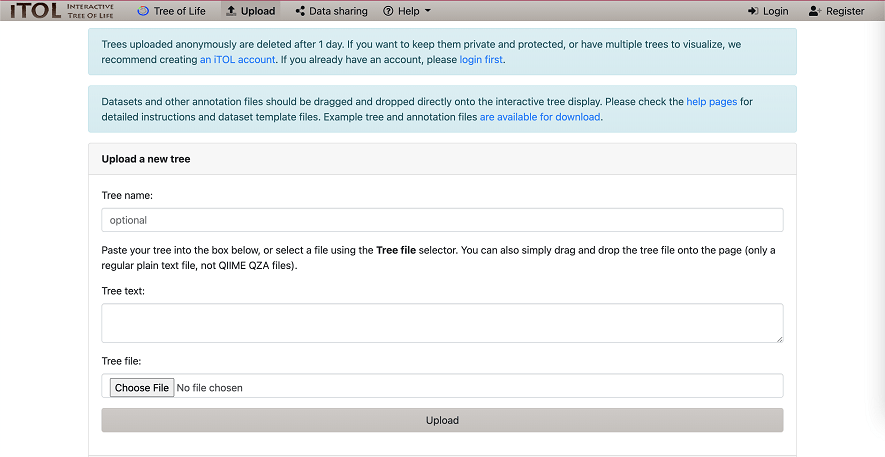
Figure 1. File Upload to the iTOL
Step 4. Evolutionary diagrams Generation
Users can adjust the parameters based on specific needs, such as display mode, label font, color, line thickness, etc.
Finally, results can be generated like demos!
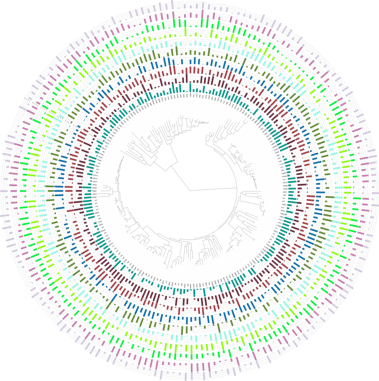
Figure 2. Bar charts datasets
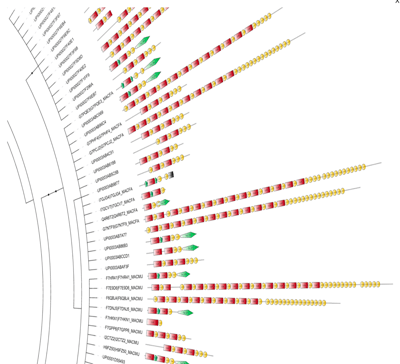
Figure 3. Protein domain architecture datasets
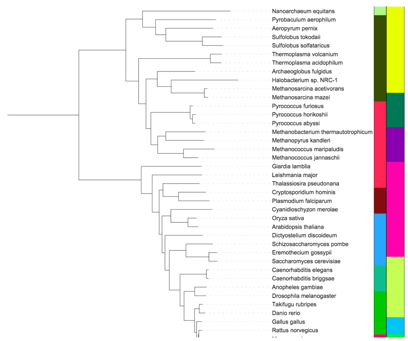
Figure 4. Color strip datasets
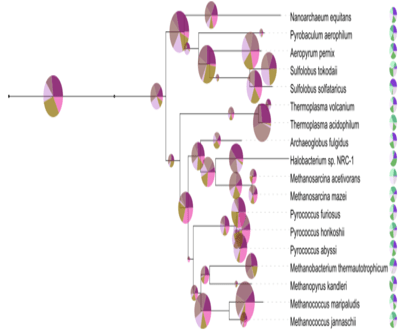
Figure 5. Pie chart datasets.
Reference
[1] Letunic, I., & Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic acids research, 47(W1), W256–W259. https://doi.org/10.1093/nar/gkz239
