Do you understand the impact of SNPs on the function of genes?
SNPs, or single nucleotide polymorphisms, are the most basic type of individual DNA variation. These straightforward modifications, which can be either transition or transversion type, happen roughly once every 1,000 (bp) throughout the whole genome. They could be in charge of interindividual variations in drug responses, diversity among individuals, genome evolution, the most prevalent familial traits like complex and common diseases like diabetes, obesity, hypertension, and psychiatric disorders. SNPs can either be silent (synonymous), change the encoded amino acids (nonsynonymous), or simply occur in non-coding regions. They may have an impact on messenger RNA (mRNA) conformation (stability), subcellular localization of mRNAs and/or proteins, promoter activity (gene expression), and thus may result in disease. [1]
Therefore, SNPs offer several opportunities for genetic, breeding, ecological, and evolutionary studies because of this. Due to their high density, scalability, and genome-wide dispersion, SNPs are regarded as suitable genomic resources in genetic research for the characterization of genetic resources and functional gene identification for features.
Let’s have a look at how different regional SNPs affect gene expression.
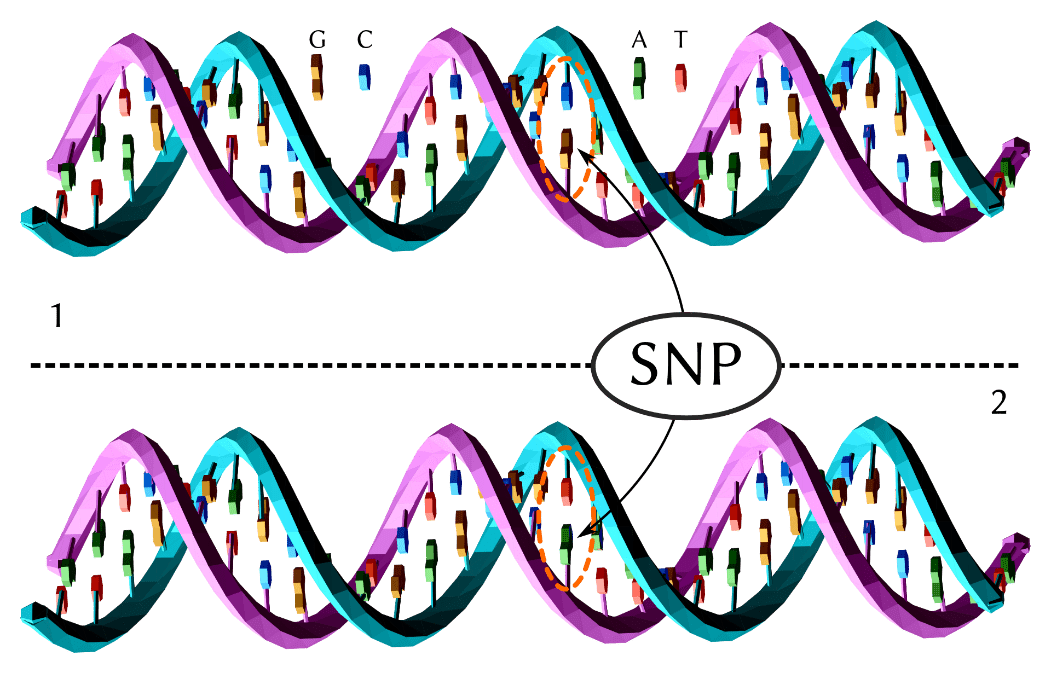
From Wikipedia: The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a G/A polymorphism)
Effect of non-synonymous coding SNPs on gene function (CDS region)
Non-synonymous coding SNPs directly change the amino acid composition of the protein encoded by the gene (cause changes at the protein level resulting from the alteration of the gene), Their function depends upon whether the variant amino acid site plays a crucial role in protein structure or function.
A protein requires a specific spatial structure to exercise its function, and the protein primary sequence directly affects the spatial structure. However, amino acids at different sites affect the structure differently, so non synonymously encoded SNPs at different positions can also have either minor or major effects on protein function. Most of the disease causing or harmful non-synonymous coding SNPs are affecting protein stability, and 1/4 of the SNPs will have an effect on protein function if they are up or down of non-synonymous coding [2].
Effect of synonymous coding SNPs on gene function
The synonymous coding SNP itself will not change the protein sequence, because not all changes in codon will result in the change of amino acid sequence, but this does not imply that this part of the SNP has no effect on the disease. There are relatively few studies on this part, but its effects are mainly from mRNA secondary structure, protein folding, and cellular localization.
This is a nonsense mutation which is a type of mutation that changes a sense codon (a codon that corresponds to one of twenty amino acids) into a chain terminating codon. Although this nonsense mutation does not change the composition of amino acids but it does change the speed of the ribosome as it passes through the mRNA fragment surrounding the SNP and the folding process in the cell which is generally considered to be synchronized with translation. This is due to the existence of codon preference in protein translation, the changing from a commonly used codon to an infrequently used codon cause the mentioned effects. These SNPs affect P-glycoprotein folding and its timing of translocation to the cell membrane, thus changing the structure of the sites of action with substrates and repressors [3].
Effect of SNPs in Intron regions on gene function
Introns make up a large proportion of DNA in eukaryotes. Although the number of SNPs distributed in the intron region are not small, but their pathogenic risk is lower than that of the SNPs in CDS and gene regulatory regions. It must be noted that the SNPs located in the first intron have a higher risk of pathogenicity than SNPs in other introns [4]d. The current study has indicated that SNPs in introns mainly rely on affecting splice site activity to influence gene function. Translation and protein sequences may be influenced by the inactivation of Splice sites.
Effect of SNPs in gene regulatory regions on gene function
Gene regulatory regions include promoter regions, enhancer regions, etc. Promoter region is a region of DNA upstream of a gene where proteins bind to initiate transcription of that gene. Enhancer Region is a short DNA region that proteins (activators) bind with to increase the likelihood of transcription of a gene.
These regions contain many regulatory elements of gene expression, such as transcription factor binding sites. The binding of these cis elements and trans elements (e.g., transcription factors) requires specific sequence composition. The changes in SNPs at these sites can alter regulatory regions binding ability to regulatory factors, which will lead to affect normal gene expression.
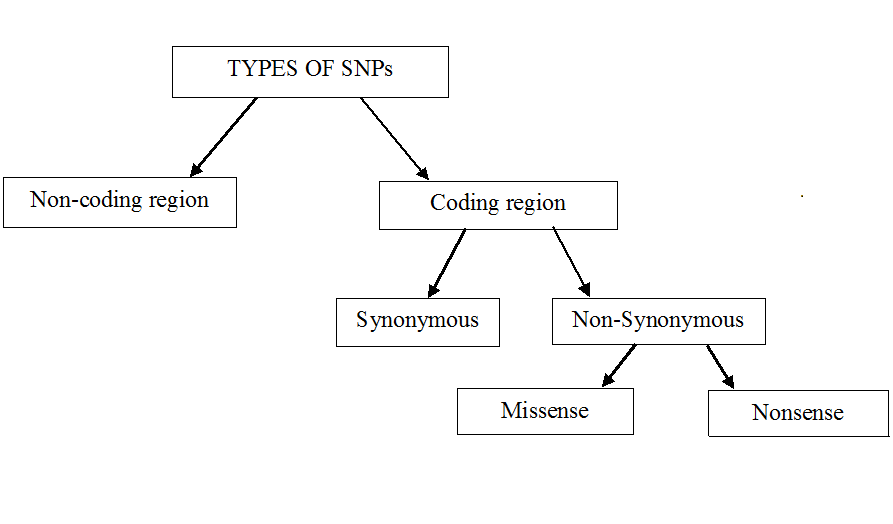
From Wikipedia: Types of single-nucleotide polymorphism (SNPs)
Reference:
- Zhao, Y., Wang, K., Wang, Wl. et al. A high-throughput SNP discovery strategy for RNA-seq data. BMC Genomics 20, 160 (2019).
- Shatoff, Elan, and Ralf Bundschuh. “Single nucleotide polymorphisms affect RNA-protein interactions at a distance through modulation of RNA secondary structures.” PLoS computational biology 16.5 (2020): e1007852.
- Komar A A . Genetics. SNPs, silent but not invisible.[J]. science, 2007, 315(5811):466-7.
- Rong Chen,Eugene V. Davydov,Marina Sirota,Atul J. Butte. Non-Synonymous and Synonymous Coding SNPs Show Similar Likelihood and Effect Size of Human Disease Association[J]. PLOS ONE,2010,5(10).
