Circular RNA: understanding its applications in therapeutics
Introduction to Circular RNA
Research surrounding ribonucleic acid (RNA) has fueled countless advancements in medical science, genomics, and biology in general. It plays a vital role in many fields of study, including disease prevention and species conservation.
Since its inception, RNA sequencing (RNA-Seq) has been pivotal in further advancing our understanding of how RNA works and its applications. These RNA studies have led to the discovery of several different types of RNA.
Unique among them is circular RNA (circRNA), a molecule that forms a closed covalent loop. This sets it apart from other linear RNA molecules, which form an open-ended chain of nucleotides.
circRNAs were first discovered in the 1970s. However, it was during the 2010s that their functions gained significant attention because of high-throughput RNA sequencing and bioinformatic analysis, especially microRNA (miRNA) sponges.
The extensive identification of circRNAs has piqued the interest of many researchers, leading to deeper investigations into their many functions. Circular RNA sequencing (circRNA-Seq) is a powerful tool that helps researchers learn more about the different functions of circRNA. In this article, we will go through the basics of the process and understand what Novogene’s circRNA sequencing services can offer.
What is circRNA, and Why is it Unique?
Circular RNAs (circRNAs) are a unique class of RNAs formed from the alternative splicing of exons from transfer RNAs (tRNAs) or pre-messenger RNAs. They can also be categorized into several types based on how they are processed:
- Exonic circRNAs
- Intronic circRNAs
- Exon-intron circRNAs
- Readthrough circRNAs
- Fusion circRNAs
- tRNA-derived circRNAs
CircRNAs are a type of non-coding RNA (ncRNA), although some circRNAs have protein-coding potential. circRNAs stand out among ncRNAs for their unique closed covalent loops, with their 5’ and 3’ ends bonded together.
Research into circRNAs has shown that they perform multiple functions:
- Transcriptional regulation
- microRNA sponging
- Translation into proteins
- Biomarkers of disease
When circRNA synthesizes with proteins, it leads to the creation of circRNA-protein complexes, which affect the organism’s life functions. As we learn more about these processes, our knowledge about how circRNA works can be used to devise novel clinical strategies.
Applications of circRNA in Therapeutics
There are many areas that can benefit from deeper circRNA research:
circRNAs are special because they have been reported to play crucial roles in various biological processes, such as regulating gene expression at the transcriptional level. They do this by interacting with gene promoters and acting as macromolecule recruiters via structure-based scaffolding. As such, they act as miRNA decoys through complementary duplex formation, as well as facilitate the translation of circRNA-specific peptides.
Riding on the heels of the development of COVID-19 vaccines by the mRNA vaccine industry, circRNA technology is also quickly emerging as a next-generation RNA therapy solution. Endogenous circRNAs are being studied to serve as new drug targets or act as disease diagnosis biomarkers. Artificially prepared circRNAs, on the other hand, can target various elements and function within cells.
Evidently, circRNA is slated to become a key technology within the global bio-economy. In addition to the COVID-19 vaccine, several new circRNA therapy enterprises have made significant breakthroughs, particularly CAR-T and protein replacement therapy. Preparing circRNA in vitro is a critical procedure that can pave the way for future advancements in clinical developments and industrialization.
circRNAs have recently gained attention in cancer research, having been identified in numerous cancer types. circRNAs have tissue-specific expression patterns, making them useful in potentially identifying disease-specific biomarkers and in executing targeted therapeutics.
Some treatments that are being studied for targeting upregulated circRNAs in cancer include RNA interference, antisense oligonucleotides, and the RNA-targeting CRISPR-Cas13 system.
In addition to this, research on circRNAs is also being explored in several areas where the application of such may be useful. Some key areas of study are:
- Identifying key characteristics of circRNAs in terms of abundance, structure, and biogenesis mechanisms
- Studying circRNA nuclear export, abundance, and turnover — and how to regulate these processes
- Exploring how circRNAs function in normal cellular processes and pathways
- Understanding the different functions of circRNAs depending on specific conditions
- Examining how circRNA is dysregulated in diseases like cancer in order to identify biomarkers and therapeutic targets
- Synthesizing artificially-engineered circRNAs with enhanced functionalities to use them for diagnostic and therapeutic purposes
CircRNA sequencing is a highly delicate and specialized process. Not very different from ncRNA-Seq, circRNA sequencing requires the added step of removing linear RNA from the library and retaining only circRNA.
The circRNA-Seq process helps researchers examine how genes are affected in various areas — such as trait expression and disease manifestation. There are four main stages to circRNA-Seq. Let’s take a closer look at each of them:
1. RNA Extraction
The first step is to extract the RNA molecules from the cells. Nucleic acids are derived and isolated from samples like individual cells, bulk tissue, and biofluids.
While DNA is found only in the cell’s nucleus, RNA exists as a single strand of nucleotides that can be found in other parts of the cell. Because it is a single strand, it is more fragile than DNA. That means extracting the RNA is a delicate process.
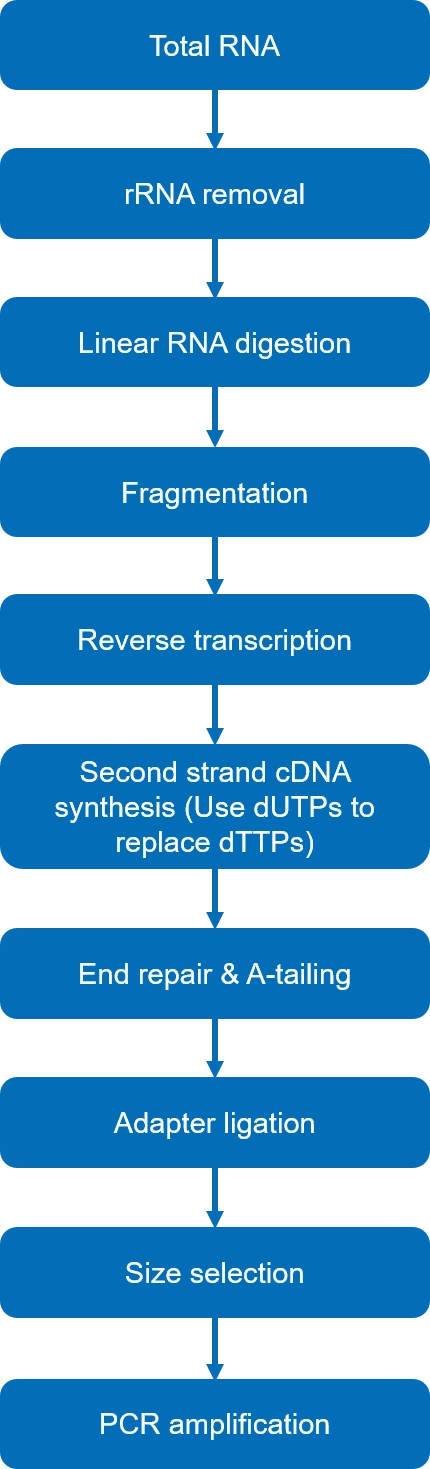
Fig 1. Library construction workflow
2. Reverse Transcription
Once the RNA has been extracted, it needs to be prepared for sequencing. Through reverse transcription, every RNA molecule is converted into a fragment of complementary DNA (cDNA). These cDNA fragments, when put together, make up the cDNA library.
For circRNA-Seq, circRNA is converted and entered into the cDNA library.
The cDNA library can then be sequenced with the aid of computers in the same way that DNA is. The sequencing can follow paired-end or single-end strategies, with the former being more expensive but significantly more detailed than the latter.
Based on the depth of the sequencing, data can be analyzed for studying genes and splicing. It helps researchers discover which genes are being transcribed by the RNA in the cell and to what degree. Bioinformatics tools are used to analyze the series of Ts, Gs, As, Cs, or reads that leave the sequencer.
Standard circRNA-Seq Analysis Pipeline
A routine circRNA-Seq project has to follow an established pipeline. For safety and accuracy, the first step is always about ensuring that the raw data matches the quality control standards. As such, the raw data is checked for error rate distribution and GC-content distribution and is passed through data filtering.
The sample’s RNA molecules are then mapped to the subject organism’s reference genome. This determines their position on the chromosome. Next is circRNA identification. Identifying the circRNA molecules helps researchers map variables like their length distribution, source genes, and location.
Once the circRNA identification is complete, researchers can predict miRNA binding sites. The total number of circRNA, both known and novel, is quantified. The final step in the circRNA-Seq pipeline is differential expression analysis, which allows the functional enrichment of circRNA source genes to be studied.
The flowchart below illustrates how the standard circRNA-Seq pipeline works:
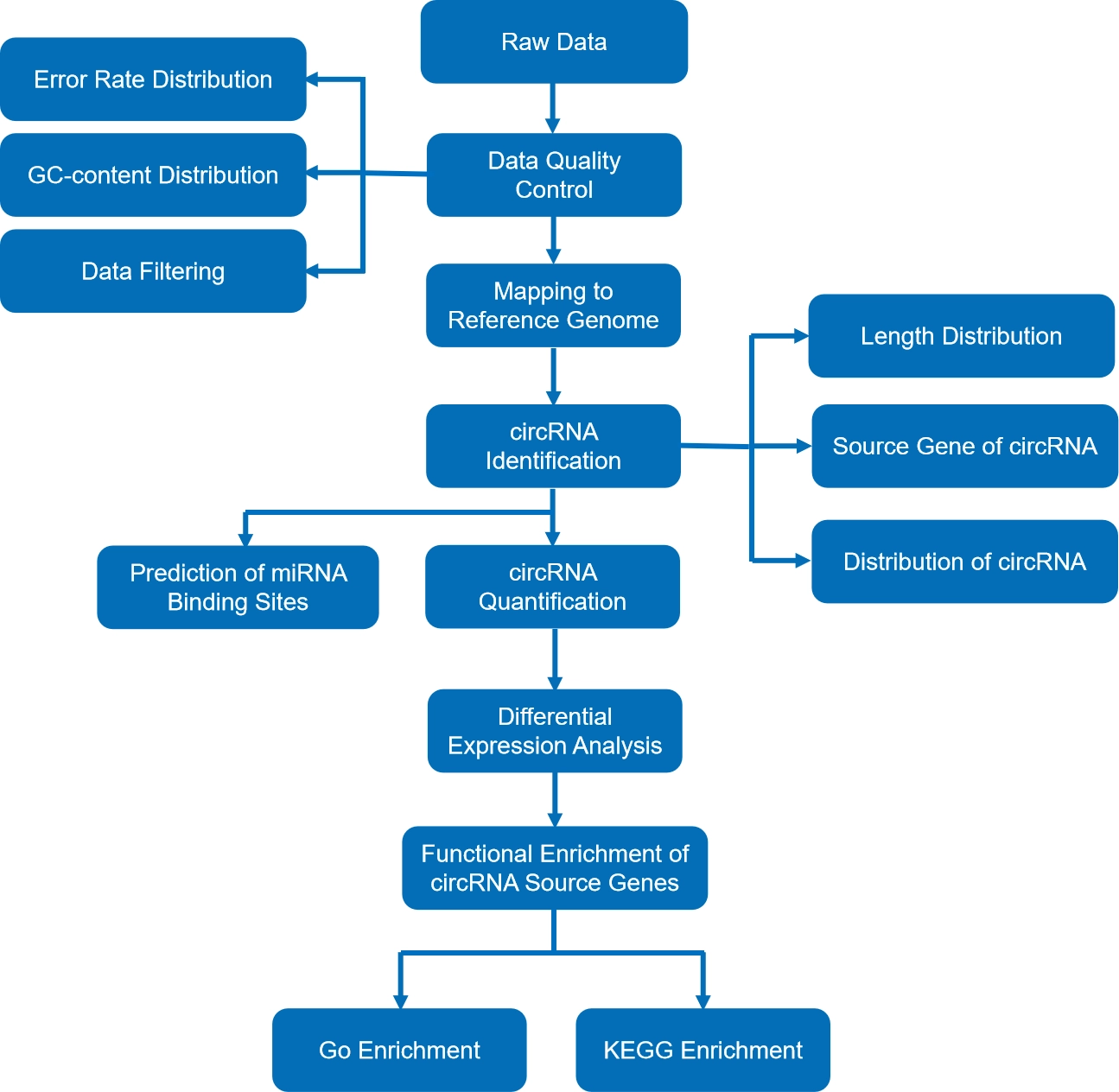
Fig 2. Bioinformatic analysis workflow
Novogene is a leading provider of genomic services and solutions with cutting-edge NGS and bioinformatics expertise. It has the largest sequencing capacity in the world, offering expert circRNA-Seq services at every stage of the process, from sample collection, RNA library preparation, and cDNA sequencing to the final bioinformatics analysis.
Novogene provides a convenient and fast solution to assist researchers in understanding the regulatory networks and the deep regulatory mechanisms of gene expression and has extensive experience in supporting many investigators in research. The service offers a comprehensive analysis performed with industry-standard software, as well as a mature in-house bioinformatics pipeline.
Novogene uses the Illumina platform to conduct its sequencing and analyze data. This process also identifies the distribution of circRNA in the cell and predicts miRNA binding sites. No matter the project, Novogene’s RNA-Seq capabilities are undoubtedly up to the task.
Qi, D., Ke, R., Huang, J. H., & Wu, E. (2023). Forging the future of circRNA therapeutics: Unleashing synthetic potential and conquering challenges. Molecular therapy. Nucleic acids, 33, 42–43. https://doi.org/10.1016/j.omtn.2023.06.002
He, A. T., Liu, J., Li, F., & Yang, B. B. (2021). Targeting circular RNAs as a therapeutic approach: current strategies and challenges. Signal transduction and targeted therapy, 6(1), 185. https://doi.org/10.1038/s41392-021-00569-5
Sun, M., & Yang, Y. (2023). Biological functions and applications of circRNAs-next generation of RNA-based therapy. Journal of molecular cell biology, 15(5), mjad031. https://doi.org/10.1093/jmcb/mjad031
Zhao, X., Zhong, Y., Wang, X., Shen, J., & An, W. (2022). Advances in Circular RNA and Its Applications. International journal of medical sciences, 19(6), 975–985. https://doi.org/10.7150/ijms.71840
Liu, X., Zhang, Y., Zhou, S., Dain, L., Mei, L., & Zhu, G. (2022). Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. Journal of controlled release : official journal of the Controlled Release Society, 348, 84–94. https://doi.org/10.1016/j.jconrel.2022.05.043
