A Beginner’s Guide to DNA Sequencing
Why do we need DNA sequencing
To comprehend the origins of life, the variations among living entities, and the course of organismal evolution. DNA sequencing could provide the underlying genetic basis (genotype) for determining what kind of cell it will become (phenotype). Each individual has a specific nucleotide base sequence that determines the characteristics of the organisms. As a result, DNA sequencing can help us to increase our understanding of the biological functions of various genes and genomes of plants, animals, and microbial communities. It provides direction to the study of recombinant DNA and to understanding the function of genes and the genome overall.
Development of Sequencing Technologies
To accomplish these goals, human research and exploration of genes and genome structure have not stopped since the invention of DNA sequence analysis technology in the 1970s. DNA sequencing techniques have advanced in recent years, generating more data and increasing sensitivity. A range of sequencing technologies are available that use different sequencing platforms, and each has different advantages.
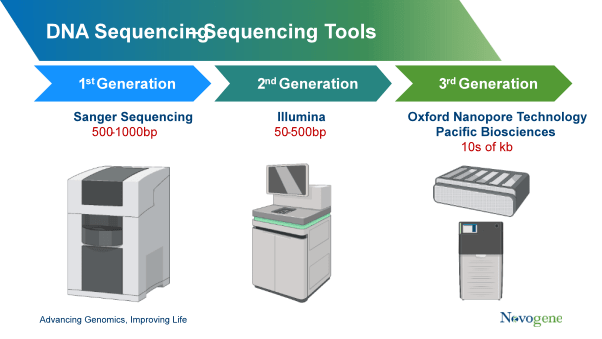
First-generation DNA sequencing tools are based on Sanger sequencing, a well-established method of determining nucleic acid sequences. It uses oligonucleotides primer to target specific DNA regions with very high accuracy and is suitable for constructing specific genomic regions (~1000 bp in length) in a larger number of samples. It is cost-efficient sequencing for single samples and simple genes. However, it has the disadvantages of slow speed, low throughput, and low sensitivity, which makes it difficult to be applied on a large-scale sequence and the low input DNA.
Second-generation DNA sequencing is represented by Illumina’s sequencing by synthesis (SBS) technology which can simultaneously sequence more data rapidly. This massively parallel sequencing approach can not only generate higher throughput and larger-scaled data but also has increased sensitivity, enabling us to go into more depth with DNA sequencing. It allows entire human genome sequencing in a thousand-dollar level. However, the limitations of next-generation sequencing are still obvious, it can’t eliminate amplification bias, and short lengths of reads are ineffective in covering the large copy number variants and structural variants.
Third-generation technologies are also based on NGS technologies and the two we will focus on here are Pacific Biosciences (PicBio) and Oxford Nanopore Technology (ONT).
PacBio offers two approaches via proprietary SMRT (Single Molecule Real Time) technology to DNA sequencing: Continuous Long Reads (CLR) and Circular Consensus Sequencing (CCS). When determining the sequence of large inserts, CLR involves the DNA polymerase only making one pass over the insert to determine the sequence. Although this method has a high error rate, it is excellent for lengthy reads. The most accurate method is CCS since it Generates multiple passes on each molecule sequenced and eventually gets high-accuracy HIFI reads. It may also be used to examine short inserts. PacBio pursues quality and uses optical signals, so it can improve the accuracy of sequencing by measuring several times, but this method has its own limitation: Expensive, Large amounts of starting material and High error rate with CLR mode.
ONT is different from all other sequencing technologies. It utilizes a nanopore method to sequence DNA and produces long reads that are longer than PacBio because it does not require a DNA polymerase chain reaction. The ultra-long read length of ONT makes it easier to detect abnormalities such as gene insertion, deletion, replication, and displacement on the sequence, providing support for cancer detection and treatment. ONT uses an electrical signal to pursue length. Each DNA base is measured at most twice. Therefore sequencing accuracy is compromised.
Applications of DNA sequencing
DNA sequencing technologies can be used to answer various research questions and the approach that you use will depend on the parameters involved in your research and the type of information you require. Here, we will give you an overview of the characteristics of each type and when it is used.
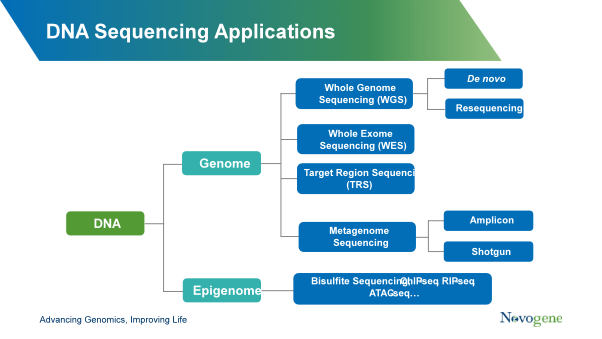
Whole genome sequencing (WGS)
WGS is used to examine all genes and non-coding DNA in a sample at a sequencing depth of >30x.
It provides a deep insight into the DNA sequence of humans, animals, plants, and microbial genomes and enables the identification and analysis of various variants of the genome.
It offers two approaches to sequencing: de novo and resequencing.
- De novo sequencing is used to assemble a genome of an organism which does not have a reference genome and generates thousands of sequencing reads.
- The resequencing approach can be used to look at an individual of a species where a reference genome exists. This technique generates millions of sequencings reads that are then aligned with the existing sequence to identify gene variations, such as insertions, deletions, single nucleotide variants, and translocation.
- Sample preparation
- Library preparation
- Sequencing
- Data Analysis
Whole Exome Sequencing (WES)
WES looks at the protein-coding regions or the exomes within the genome and aims to identify genetic variants that alter protein function and is a useful tool for identifying disease-related mutations associated with Mendelian disorders. WES is more accurate than WGS and has a minimum sequencing depth of 50 – 100x. WES is cost-effective compared to WGS and can be applied in both academic research and clinical diagnostics. It helps to identify genetic variants that alter protein sequences, including those responsible for Mendelian and poly genic diseases, such as Alzheimer’s disease.
Target Region Sequencing (TRS)
TRS is used to sequence a select set of genes or regions that have a specific function, for example, they may be linked with a specific disease or a certain phenotype. There are two types of TRS: hybridization capture and amplicon sequencing, with hybridization capture being used to examine larger gene content.
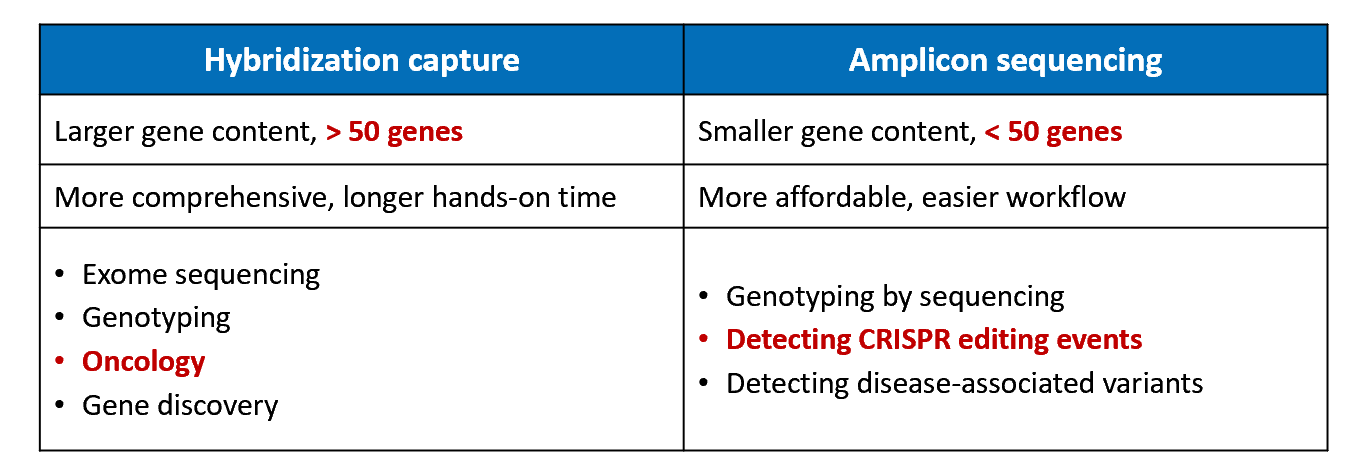
These approaches can look at 100 – 500 genes and the average sequencing depth is great than 500x. TRS has many applications including cancer research, human population studies, and the discovery of biomarkers and therapeutic targets.
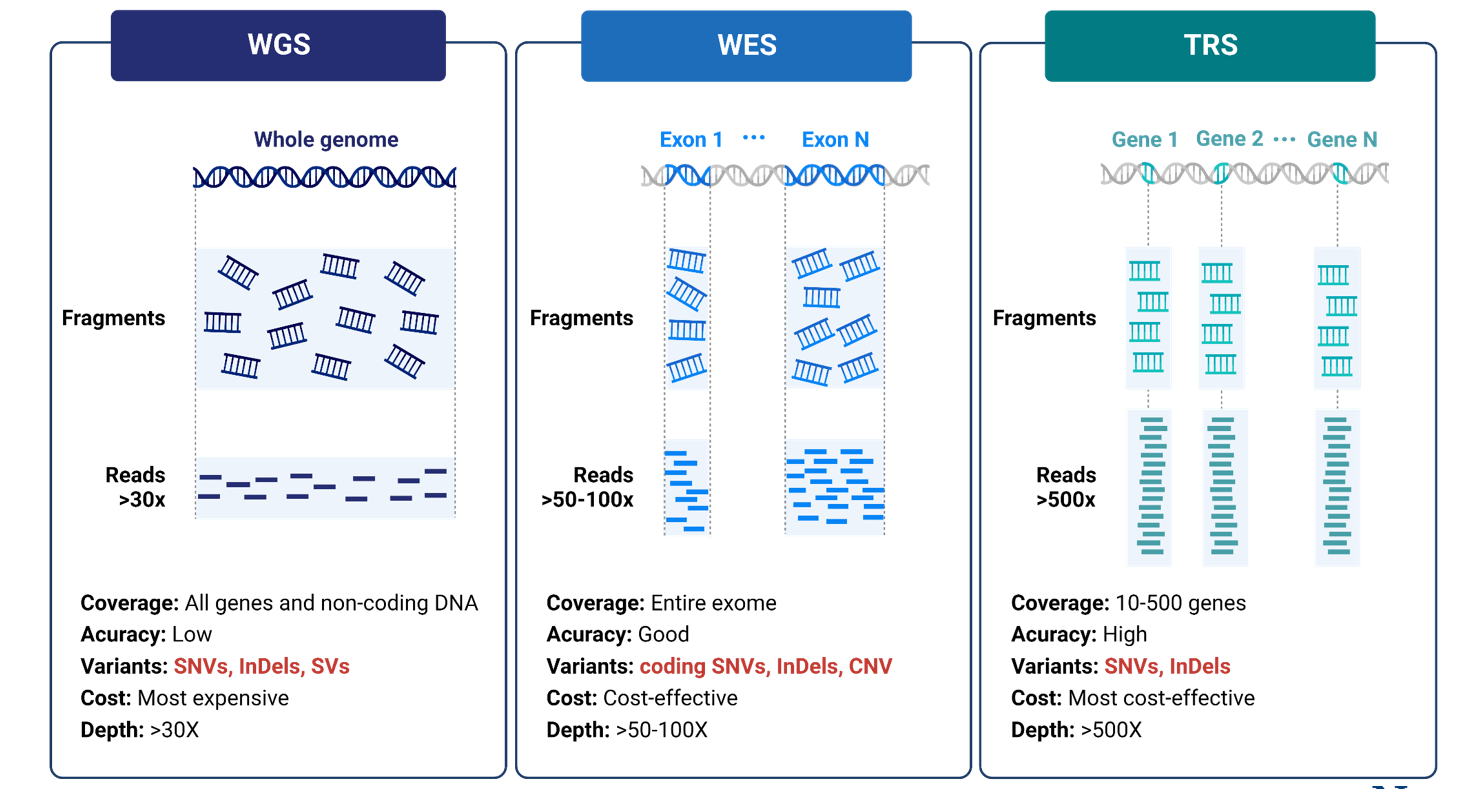
The comparison of three major technology.
Metagenomics
Metagenomic sequencing is used to characterize microbial communities, providing a comprehensive insight into the composition of and diversity within these communities and any interactions that occur. There are two approaches to metagenomic sequencing: amplicon sequencing and shotgun sequencing.
Epigenetics
Epigenetics is the study of changes that occur in the phenotype of the gene as a result of the environment without changing the genotype. Several different techniques can be used depending on your question, including ChIP-seq, which is used to examine histone modifications and transcription factors; Hi-C sequencing, which can be used to look at chromatin conformation; and Whole-genome bisulphite sequencing, which targets DNA methylation.
DNA Sequencing Requirements and Workflow
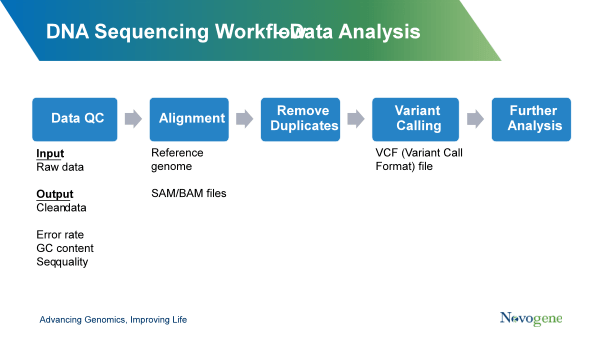
The workflow for all these techniques is relatively straightforward and involves four steps:
The samples required will depend on the type of sequencing being used. Good quality DNA is vital to achieving good sequencing results, so all samples provided will first go through quality control (QC) to ensure that they are of a high enough quality for sequencing.
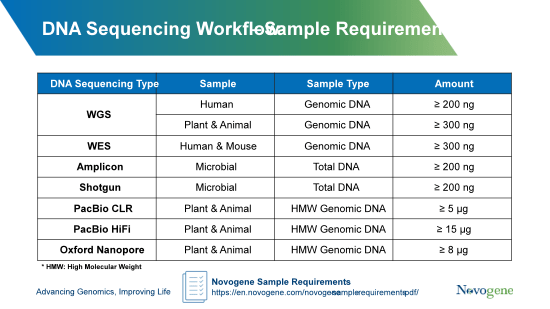
Once the DNA has passed QC, then library preparation will be carried out based on the requirements of the sequencing method you are using. Post-sequencing, the sequencing reads will go through QC to ensure sequence quality and remove any low-quality sequences or those with high error rates. Depending on your requirements, the sequences can then be aligned to the reference genome and any duplicates removed before further analysis is carried out.
Novogene is a leading provider of genomic services and solutions with cutting-edge NGS and bioinformatics expertise and the largest sequencing capacity in the world. Precisely because our supercomputing capability allows us to analyse 280,000 whole human genomes per month, this capability aligns with our unsurpassed NGS capacity, enabling us to run many projects of different sizes and still deliver the data to our customers in a timely fashion. About DNA sequencing program, we offer a range of DNA sequencing services on platforms outlined above and provide WGS, WES, and TRS sequencing, as well as metagenomics and more specific sequencing approaches such as Chip-seq. Almost all of your demands for DNA sequencing can be met by that.
For more information on DNA sequencing advances and the services Novogene offers, listen to our webinar available here: A Beginner’s Guide to DNA Sequencing
