Novogene Unravels the First Soybean Pan-genome
Experts from Novogene, Chinese Academy of Agricultural Sciences, Peking University and other institutes have identified the soybean genetic diversity and main agronomic gene using pan-genome analysis. This study published in Nature Biotechnology (IF: 39.08) facilitates the harnessing of untapped genetic diversity from wild soybean for enhancement of elite cultivars.
Abstract (De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits):
Wild relatives of crops are an important source of genetic diversity for agriculture, but their gene repertoire remains largely unexplored. We report the establishment and analysis of a pan-genome of Glycine soja, the wild relative of cultivated soybean Glycine max, by sequencing and de novo assembly of seven phylogenetically and geographically representative accessions. Intergenomic comparisons identified lineage-specific genes and genes with copy number variation or large-effect mutations, some of which show evidence of positive selection and may contribute to variation of agronomic traits such as biotic resistance, seed composition, flowering and maturity time, organ size and final biomass. Approximately 80% of the pan-genome was present in all seven accessions (core), whereas the rest was dispensable and exhibited greater variation than the core genome, perhaps reflecting a role in adaptation to diverse environments.
For more information please visit the original link:
https://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.2979.html
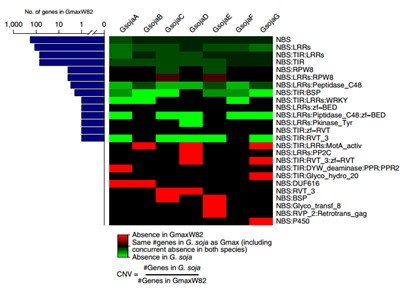
Figure Heatmap for CNV of resistance-related PFAM gene categories across all accessions
