NovaSeq X Plus 25B at Novogene: Redefining Genomic Sequencing Excellence
Technical Breakdown and Exclusive First Look: Unmatched Precision in Sequencing
Introduction
We are thrilled to introduce the NovaSeq X Plus 25B flow cell (FC), a state-of-the-art sequencing platform with unmatched throughput capabilities. The 25B flow cell enables you to step into the future of genomics with Illumina’s latest innovation. This remarkable high-density 25B flow cell empowers your research with unparalleled capabilities.
Unprecedented High Throughput Potential of the 25B FC
The NovaSeq X Plus System, loaded with 25B FC offers the unmatched throughput and accuracy needed to enable more data-intensive applications and deliver the most cost-effective solution. The NovaSeq X Plus is Illumina’s most powerful sequencing system yet, capable of generating up to 16 terabases (TB) of output (or up to 52 billion single reads) per dual 25B flow cell run. This represents a remarkable 2.5x increase from the 10B FC and an incredible 3x increase from the S4 FC (Table 1).
The performance of the NovaSeq X Series reduces the cost per gigabase (Gb) by up to 60% compared to the NovaSeq 6000 System [1]. With the cutting-edge NovaSeq X Plus 25B sequencing, you can enjoy the most cost-effective sequencing service for your research.
| NovaSeq X Plus 25B | NovaSeq X Plus 10B | NovaSeq 6000 S4 | |
| Flow Cells per Run | 2 | 2 | 2 |
| Lanes per FC | 8 | 8 | 4 |
| Sequencing Mode Available at Novogene | PE150 | ||
| Theoretical Data Output per Lane (PE 150) | 1000 Gb3.3B paired reads | 375 Gb1.25B paired reads | 800 Gb 2.7B paired reads |
| Chemistry for Patterned Flow Cells | XLEAP-SBS Chemistry 2x faster cycle time 3x accuracy increase [1] | SBS Chemistry | |
| Channel Chemistry | 2-channel chemistry (blue-green optics) | 2-channel chemistry (red-green optics) | |
| Quality at PE150 | ≥ 85% of bases higher than Q30 | ≥ 85% of bases higher than Q30 | |
| Acceptable Premade Library Type | Broad library types with Illumina compatibility | ||
Table 1: Technical Comparison of Sequencing Platforms: NovaSeq X Plus 10B vs. NovaSeq X Plus 25B vs. NovaSeq 6000 S4
Unmatched Precision with 25B
In our initial evaluation using a single 25B FC with diverse in-house and client library samples, we observed remarkable data output surpassing 10T, averaging 1200 Gb per lane (Table 2). Notably, the majority of in-house library samples showcased exceptional data quality, with approximately 96% of bases achieving quality scores above Q30. For comparison, both NovaSeq 6000 S4 and X Plus have an established Q30 benchmark of 85%.
| Type | Lane 1 | Lane 2 | Lane 3 | Lane 4 | Lane 5 | Lane 6 | Lane 7 | Lane 8 |
|
Library Type |
WES Metagenomic WGS ChIP mRNA Strand-specific mRNA Metatranscriptome lncRNA circRNA |
WES Metagenomic WGS ChIP mRNA Strand-specific mRNA Metatranscriptome lncRNA circRNA |
Client Plant & Animal WGS |
Client Single Cell RNA |
WGBS PCR-Free Panel |
WGBS PCR-Free Panel |
CUT&TAG ATAC Ribo RIP RNA methyl UMI sRNA UMI mRNA Hi-C Exosome lncRNA 10x 3′ 10x 5′ |
CUT&TAG ATAC Ribo RIP RNA methyl UMI sRNA UMI mRNA Hi-C Exosome lncRNA 10x 3′ 10x 5′ |
| Data Output / Lane (Gb) | 1279 | 1280 | 1309 | 1198 | 1344 | 1359 | 1300 | 1303 |
| Data Output/Lane (Gb) (excluding index reads) | 1206 | 1208 | 1235 | 1130 | 1268 | 1282 | 1227 | 1230 |
| Q30 | 95.85% | 95.77% | 94.08% | 77.99% | 95.55% | 95.89% | 91.7% | 91.67% |
Table 2: Library Pooling and Configuration for 25B Flow Cell Evaluation−−A single 25B FC was tested using a diverse range of library types
WES: Whole Exome Sequencing; Metagenomic: Metagenomic Library; WGS: Whole Genome Sequencing; ChIP: Chromatin Immunoprecipitation Sequencing; mRNA: Eukaryotic mRNA Sequencing; Metatranscriptome: Metatranscriptomic Library; LncRNA: Long Noncoding RNA Sequencing; circRNA: Circular RNA Sequencing; WGBS: Whole Genome Bisulfite Sequencing; PCR-free: PCR-free WGS; Panel: Gene Panel Sequencing; CUT&TAG: Cleavage Under Targets and Tagmentation; ATAC: ATAC-seq; Ribo: Ribosome Profiling; RIP: RNA Immunoprecipitation Sequencing; RNA methyl: RNA Methylation Sequencing; UMI sRNA: UMI Small RNA-seq; UMI mRNA: UMI mRNA-seq; Hi-C: Hi-C seq; 10x 3′: 10x Genomics Chromium Single Cell 3′ Gene Expression; 10x 5′: 10x Genomics Chromium Single Cell 5′ Gene Expression.
Platform Variations Decoded
We are pleased to present a comprehensive data analysis encompassing our principal sequencing products: Human Whole Genome Sequencing (WGS), Whole Exome Sequencing (WES), Human mRNA Sequencing, Metagenomics Library Sequencing, and Whole Genome Bisulfite Sequencing (WGBS). Our rigorous evaluation involved testing identical samples across three distinct configurations: NovaSeq 6000 S4, NovaSeq X Plus 10B, and NovaSeq X Plus 25B, facilitating a robust comparative assessment of performance across these sequencing methodologies. To access and download the full text analysis and report, please find the link at the bottom of the article.
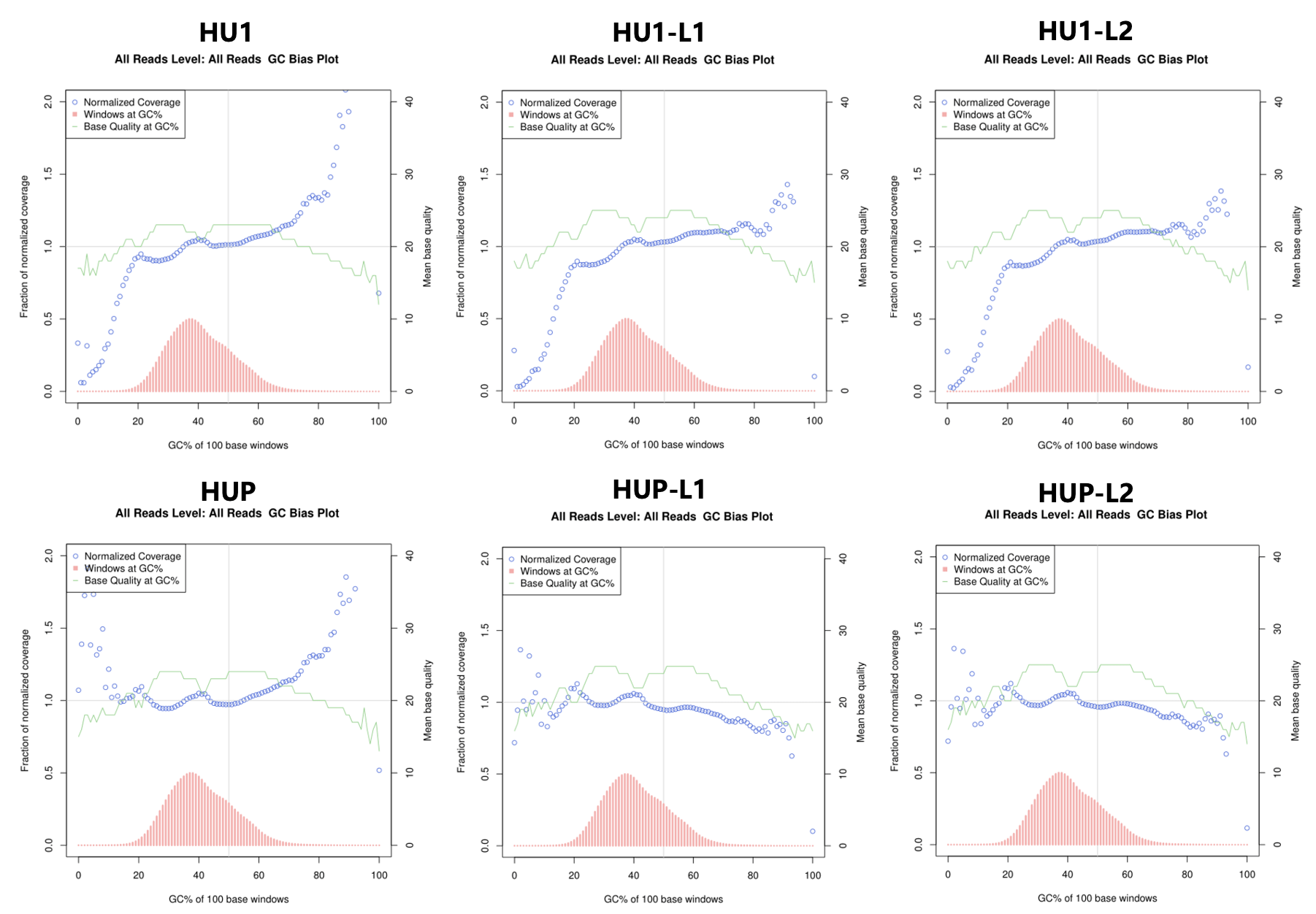
Figure: GC Bias Plots of Human WGS Samples
HU1: Control Sample; HU1-L1 and HU1-L2: Human WGS Samples on 25B Lane 1 or Lane 2; HUP: Control PCR-free Sample; HUP1-L1 and
HUP1-L2: Human PCR-free WGS Samples on 25B Lane 1 or Lane 2.
Reference
[1] Illumina. NovaSeq X and X Plus System specification sheet. https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/novaseq-x-series-spec-sheet-m-us-00197/novaseq-x-series-specification-sheet-m-us-00197.pdf
[2] ENCODE. Whole-Genome Bisulfite Sequencing Data Standards and Processing Pipeline. https://www.encodeproject.org/data-standards/wgbs/
Related Links
Comprehensive 25B Test Report with Supplementary Figures
Early Bird Special: NovaSeg X Plus 25B for Pre-made Library Sequencing Service
