Another breakthrough: Genome of allotetraploid cotton has been decoded
Collaborated with Nanjing Agriculture University and University of Texas, experts from Novogene decoded the genome of allotetraploid cotton. The study, which will provide a resource for fiber improvement, is just published in Nature Biotechnology (IF: 39.08).
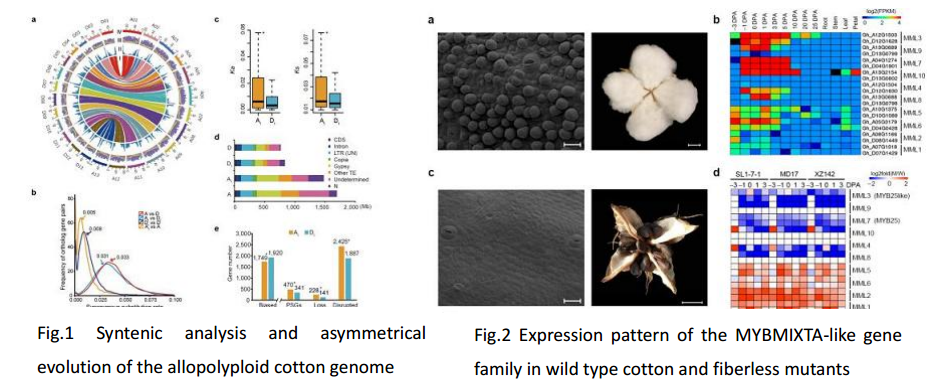
Abstract (Sequencing of allotetraploid cotton (Gossypiumhirsutum L. acc. TM-1) provides a resource for fiber improvement)
Upland cotton is a model for polyploid crop domestication and transgenic improvement. Here we sequenced the allotetraploid Gossypiumhirsutum L. acc. TM-1 genome by integrating whole-genome shotgun reads, bacterial artificial chromosome (BAC)-end sequences and genotype-by-sequencing genetic maps. We assembled and annotated 32,032 A-subgenome genes and 34,402 D-subgenome genes. Structural rearrangements, gene loss, disrupted genes and sequence divergence were more common in the A subgenome than in the D subgenome, suggesting asymmetric evolution. However, no genome-wide expression dominance was found between the subgenomes. Genomic signatures of selection and domestication are associated with positively selected genes (PSGs) for fiber improvement in the A subgenome and for stress tolerance in the D subgenome. This draft genome sequence provides a resource for engineering superior cotton lines.
For more information please visit the original link:
https://www.nature.com/nbt/journal/vaop/ncurrent/full/nbt.3207.html